3CSH
 
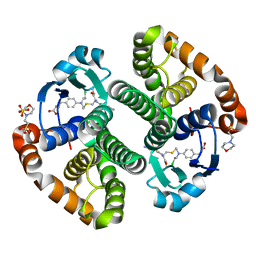 | |
3D1A
 
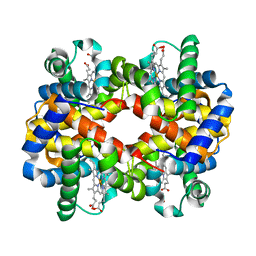 | |
4B7T
 
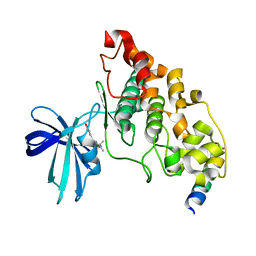 | | Glycogen Synthase Kinase 3 Beta complexed with Axin Peptide and Leucettine L4 | | Descriptor: | (5Z)-5-(1,3-benzodioxol-5-ylmethylidene)-3-methyl-2-(propan-2-ylamino)imidazol-4-one, AXIN-1, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Oberholzer, A.E, Pearl, L.H. | | Deposit date: | 2012-08-22 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.772 Å) | | Cite: | Selectivity, Cocrystal Structures, and Neuroprotective Properties of Leucettines, a Family of Protein Kinase Inhibitors Derived from the Marine Sponge Alkaloid Leucettamine B.
J.Med.Chem., 55, 2012
|
|
2PCC
 
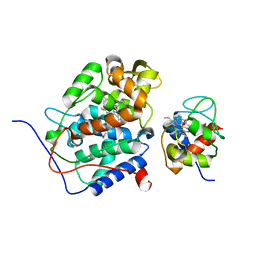 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN ELECTRON TRANSFER PARTNERS, CYTOCHROME C PEROXIDASE AND CYTOCHROME C | | Descriptor: | CYTOCHROME C PEROXIDASE, ISO-1-CYTOCHROME C, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pelletier, H, Kraut, J. | | Deposit date: | 1993-04-14 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a complex between electron transfer partners, cytochrome c peroxidase and cytochrome c.
Science, 258, 1992
|
|
6Y6B
 
 | | Crystal structure of human 14-3-3 gamma in complex with CaMKK2 14-3-3 binding motif Ser100 and 16-OMe-Fusicoccin H | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[[(1~{S},3~{R},6~{S},7~{S},8~{R},9~{R},10~{R},11~{R},14~{S})-14-(me thoxymethyl)-3,10-dimethyl-9-oxidanyl-6-propan-2-yl-8-tricyclo[9.3.0.0^{3,7}]tetradecanyl]oxy]oxane-3,4,5-triol, 14-3-3 protein gamma, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Lentini Santo, D, Obsilova, V, Obsil, T. | | Deposit date: | 2020-02-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Stabilization of Protein-Protein Interactions between CaMKK2 and 14-3-3 by Fusicoccins.
Acs Chem.Biol., 15, 2020
|
|
4X7L
 
 | | Co-crystal Structure of PERK bound to 4-{2-amino-4-methyl-3-[2-(methylamino)-1,3-benzothiazol-6-yl]benzoyl}-1-methyl-2,5-diphenyl-1,2-dihydro-3H-pyrazol-3-one inhibitor | | Descriptor: | 4-{2-amino-4-methyl-3-[2-(methylamino)-1,3-benzothiazol-6-yl]benzoyl}-1-methyl-2,5-diphenyl-1,2-dihydro-3H-pyrazol-3-one, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3, GLYCEROL, ... | | Authors: | Shaffer, P.L, Long, A.M, Chen, H. | | Deposit date: | 2014-12-09 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of 1H-Pyrazol-3(2H)-ones as Potent and Selective Inhibitors of Protein Kinase R-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 58, 2015
|
|
8HOH
 
 | | Crystal structure of Bcl-2 G101V in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
7OBR
 
 | | RNC-SRP early complex | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Ban, N. | | Deposit date: | 2021-04-23 | | Release date: | 2021-07-21 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle.
Cell Rep, 36, 2021
|
|
1SR0
 
 | | Crystal structure of signalling protein from sheep(SPS-40) at 3.0A resolution using crystal grown in the presence of polysaccharides | | Descriptor: | beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, signal processing protein | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Singh, N, Kumar, J, Sharma, S, Singh, T.P. | | Deposit date: | 2004-03-22 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of signalling protein from sheep(SPS-40) at 3.0A resolution using crystal grown in the presence of polysaccharides
To be Published
|
|
3ECA
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI L-ASPARAGINASE, AN ENZYME USED IN CANCER THERAPY (ELSPAR) | | Descriptor: | ASPARTIC ACID, L-asparaginase 2 | | Authors: | Swain, A.L, Jaskolski, M, Housset, D, Rao, J.K.M, Wlodawer, A. | | Deposit date: | 1993-07-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Escherichia coli L-asparaginase, an enzyme used in cancer therapy.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
6NE0
 
 | | Structure of double-stranded target DNA engaged Csy complex from Pseudomonas aeruginosa (PA-14) | | Descriptor: | CRISPR RNA (60-MER), CRISPR target DNA (44-MER), CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chowdhury, S, Rollins, M.F, Carter, J, Golden, S.M, Miettinen, H.M, Santiago-Frangos, A, Faith, D, Lawrence, M.C, Wiedenheft, B, Lander, G.C. | | Deposit date: | 2018-12-15 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry.
Mol. Cell, 74, 2019
|
|
1IRV
 
 | |
8HAD
 
 | |
8HAC
 
 | |
1IMQ
 
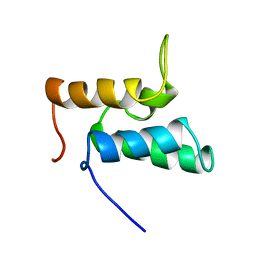 | | COLICIN E9 IMMUNITY PROTEIN IM9, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | IM9 | | Authors: | Osborne, M.J, Breeze, A.L, Lian, L.Y, Reilly, A, James, R, Kleanthous, C, Moore, G.R. | | Deposit date: | 1996-05-30 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and 13C nuclear magnetic resonance assignments of the colicin E9 immunity protein Im9.
Biochemistry, 35, 1996
|
|
7O06
 
 | |
1IVC
 
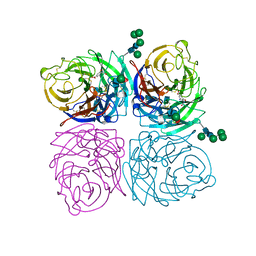 | | STRUCTURES OF AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(ACETYLAMINO)-5-AMINO-3-HYDROXYBENZOIC ACID, CALCIUM ION, ... | | Authors: | Jedrzejas, M.J, Luo, M. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of aromatic inhibitors of influenza virus neuraminidase.
Biochemistry, 34, 1995
|
|
6ALI
 
 | |
7O0S
 
 | |
7O3B
 
 | |
7S25
 
 | | ROCK1 IN COMPLEX WITH LIGAND G4998 | | Descriptor: | 2-[3-(methoxymethyl)phenyl]-N-[4-(1H-pyrazol-4-yl)phenyl]acetamide, CHLORIDE ION, Rho-associated protein kinase 1 | | Authors: | Ganichkin, O, Harris, S.F, Steinbacher, S. | | Deposit date: | 2021-09-03 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.337 Å) | | Cite: | Chemical space docking enables large-scale structure-based virtual screening to discover ROCK1 kinase inhibitors.
Nat Commun, 13, 2022
|
|
7S26
 
 | | ROCK1 IN COMPLEX WITH LIGAND G5018 | | Descriptor: | 2-[methyl(phenyl)amino]-1-[4-(1H-pyrrolo[2,3-b]pyridin-3-yl)-3,6-dihydropyridin-1(2H)-yl]ethan-1-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Rho-associated protein kinase 1 | | Authors: | Ganichkin, O, Harris, S.F, Steinbacher, S. | | Deposit date: | 2021-09-03 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Chemical space docking enables large-scale structure-based virtual screening to discover ROCK1 kinase inhibitors.
Nat Commun, 13, 2022
|
|
2J11
 
 | | p53 tetramerization domain mutant Y327S T329G Q331G | | Descriptor: | CELLULAR TUMOR ANTIGEN P53 | | Authors: | Carbajo, R.J, Mora, P, Sanchez del Pino, M.M, Perez-Paya, E, Pineda-Lucena, A. | | Deposit date: | 2006-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solvent-exposed residues located in the beta-sheet modulate the stability of the tetramerization domain of p53--a structural and combinatorial approach.
Proteins, 71, 2008
|
|
1ILD
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI N156A ACTIVE SITE MUTANT pH 4.6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, OUTER MEMBRANE PHOSPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Van Eerde, J.H, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2001-05-08 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn-His interaction in the catalytic triad.
Protein Sci., 10, 2001
|
|
1IM0
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI N156A ACTIVE SITE MUTANT PH 8.3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, OUTER MEMBRANE PHSOPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Van Eerde, J.H, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2001-05-09 | | Release date: | 2001-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn-His interaction in the catalytic triad.
Protein Sci., 10, 2001
|
|
