4E37
 
 | | Crystal Structure of P. aeruginosa catalase, KatA tetramer | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | VanderWielen, B.D, Wilson, J.J, Kovall, R.A. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | KatA, the Major Catalase in Pseudomonas aeruginosa, Assists in Protecting Anaerobic Bacteria From Metabolic and Exogenous Nitric Oxide
To be Published
|
|
8IN8
 
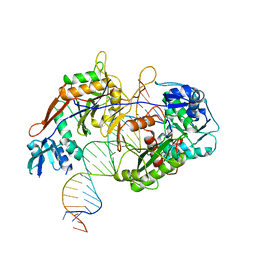 | | Cryo-EM structure of the target ssDNA-bound SIR2-APAZ/Ago-gRNA quaternary complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*CP*GP*TP*CP*TP*AP*AP*GP*AP*AP*AP*CP*CP*AP*TP*TP*AP*A)-3'), MAGNESIUM ION, Piwi domain protein, ... | | Authors: | Zhang, H, Li, Z, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-03-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
3CLC
 
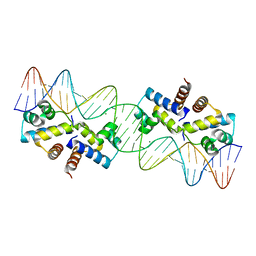 | | Crystal Structure of the Restriction-Modification Controller Protein C.Esp1396I Tetramer in Complex with its Natural 35 Base-Pair Operator | | Descriptor: | 35-MER, MAGNESIUM ION, Regulatory protein | | Authors: | McGeehan, J.E, Streeter, S.D, Thresh, S.J, Ball, N, Ravelli, R.B, Kneale, G.G. | | Deposit date: | 2008-03-18 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of the genetic switch that regulates the expression of restriction-modification genes.
Nucleic Acids Res., 36, 2008
|
|
4W61
 
 | |
4MER
 
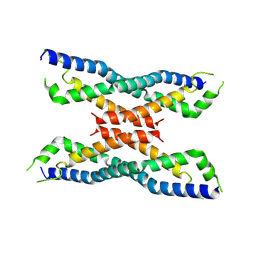 | | Crystal structure of the novel protein and virulence factor sHIP (Q99XU0) from Streptococcus pyogenes | | Descriptor: | streptococcal Histidine-rich glycoprotein Interacting Protein | | Authors: | Wisniewska, M, Happonen, L, Frick, M.-I, Bjorck, L, Streicher, W, Malmstrom, J, Wikstrom, M. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-21 | | Last modified: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Functional and Structural Properties of a Novel Protein and Virulence Factor (Protein sHIP) in Streptococcus pyogenes.
J.Biol.Chem., 289, 2014
|
|
4HKA
 
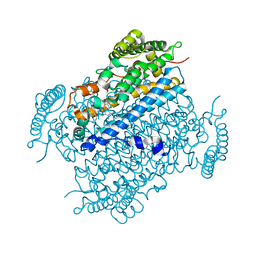 | |
4WBC
 
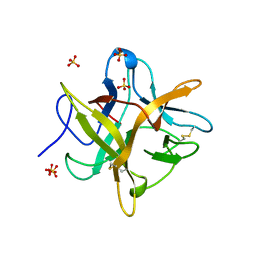 | | 2.13 A STRUCTURE OF A KUNITZ-TYPE WINGED BEAN CHYMOTRYPSIN INHIBITOR PROTEIN | | Descriptor: | PROTEIN (CHYMOTRYPSIN INHIBITOR), SULFATE ION | | Authors: | Ravichandran, S, Sen, U, Chakrabarti, C, Dattagupta, J.K. | | Deposit date: | 1999-03-04 | | Release date: | 1999-03-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.138 Å) | | Cite: | Cryocrystallography of a Kunitz-type serine protease inhibitor: the 90 K structure of winged bean chymotrypsin inhibitor (WCI) at 2.13 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
8ITE
 
 | |
3D8T
 
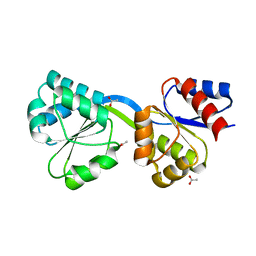 | | Thermus thermophilus Uroporphyrinogen III Synthase | | Descriptor: | ACETATE ION, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
8IA9
 
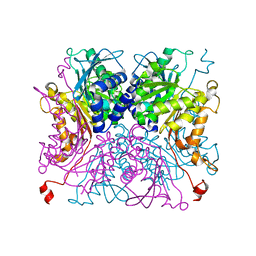 | |
3D8S
 
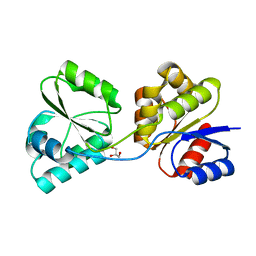 | | Thermus thermophilus Uroporphyrinogen III Synthase | | Descriptor: | GLYCEROL, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
8IAA
 
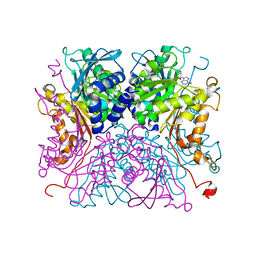 | |
4UZB
 
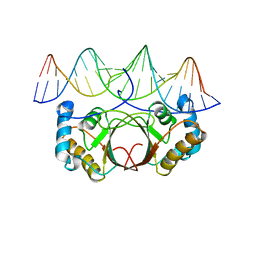 | | KSHV LANA (ORF73) C-terminal domain mutant bound to LBS1 DNA (R1039Q, R1040Q, K1055E, K1109A, D1110A, A1121E, K1138S, K1140D, K1141D) | | Descriptor: | LANA BINDING SITE 1 DNA, ORF 73 | | Authors: | Hellert, J, Krausze, J, Luhrs, T. | | Deposit date: | 2014-09-05 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.865 Å) | | Cite: | The 3D Structure of Kaposi Sarcoma Herpesvirus Lana C-Terminal Domain Bound to DNA.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4URF
 
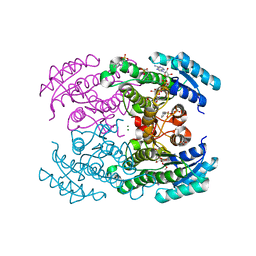 | | Molecular Genetic and Crystal Structural Analysis of 1-(4- Hydroxyphenyl)-Ethanol Dehydrogenase from Aromatoleum aromaticum EbN1 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ACETATE ION, BICARBONATE ION, ... | | Authors: | Buesing, I, Hoeffken, H.W, Breuer, M, Woehlbrand, L, Hauer, B, Rabus, R. | | Deposit date: | 2014-06-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular Genetic and Crystal Structural Analysis of 1-(4-Hydroxyphenyl)-Ethanol Dehydrogenase from 'Aromatoleum Aromaticum' Ebn1.
J.Mol.Microbiol.Biotechnol., 25, 2015
|
|
4MS2
 
 | | Structural basis of Ca2+ selectivity of a voltage-gated calcium channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-18 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
4V40
 
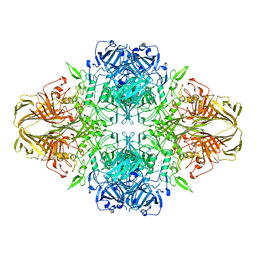 | | BETA-GALACTOSIDASE | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Jacobson, R.H, Zhang, X, Dubose, R.F, Matthews, B.W. | | Deposit date: | 1994-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of beta-galactosidase from E. coli.
Nature, 369, 1994
|
|
4MTF
 
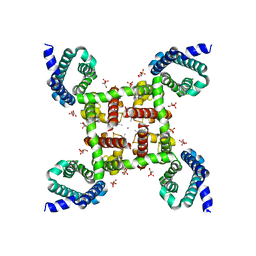 | | Structural Basis of Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-19 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
3EED
 
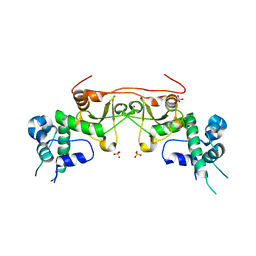 | | Crystal structure of human protein kinase CK2 regulatory subunit (CK2beta; mutant 1-193) | | Descriptor: | Casein kinase II subunit beta, SULFATE ION, ZINC ION | | Authors: | Niefind, K, Raaf, J, Issinger, O.-G. | | Deposit date: | 2008-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The interaction of CK2{alpha} and CK2{beta}, the subunits of protein kinase CK2, requires CK2{beta} in a preformed conformation and is enthalpically driven
Protein Sci., 17, 2008
|
|
4WID
 
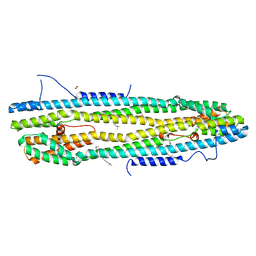 | | Crystal structure of the immediate-early 1 protein (IE1) at 2.31 angstrom (tetragonal form after crystal dehydration) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, RhUL123 | | Authors: | Klingl, S, Scherer, M, Sevvana, M, Muller, Y.A, Stamminger, T. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Cytomegalovirus IE1 Protein Reveals Targeting of TRIM Family Member PML via Coiled-Coil Interactions.
Plos Pathog., 10, 2014
|
|
4CSK
 
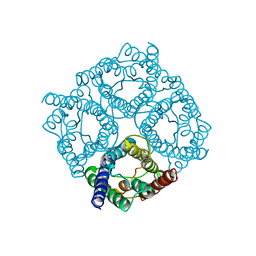 | | human Aquaporin | | Descriptor: | AQUAPORIN-1 | | Authors: | Ruiz-Carrillo, D, To-Yiu-Ying, J, Darwis, D, Soon, C.H, Cornvik, T, Torres, J, Lescar, J. | | Deposit date: | 2014-03-08 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Crystallization and Preliminary Crystallographic Analysis of Human Aquaporin 1 at a Resolution of 3.28 A.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4CMR
 
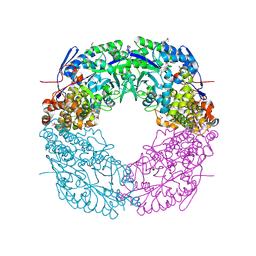 | | The crystal structure of novel exo-type maltose-forming amylase(Py04_0872) from Pyrococcus sp. ST04 | | Descriptor: | GLYCOSYL HYDROLASE/DEACETYLASE FAMILY PROTEIN | | Authors: | Park, K.-H, Jung, J.-H, Park, C.-S, Woo, E.-J. | | Deposit date: | 2014-01-17 | | Release date: | 2014-10-22 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Features Underlying the Selective Cleavage of a Novel Exo-Type Maltose-Forming Amylase from Pyrococcus Sp. St04
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1F93
 
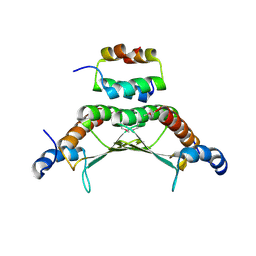 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN THE DIMERIZATION DOMAIN OF HNF-1 ALPHA AND THE COACTIVATOR DCOH | | Descriptor: | DIMERIZATION COFACTOR OF HEPATOCYTE NUCLEAR FACTOR 1-ALPHA, HEPATOCYTE NUCLEAR FACTOR 1-ALPHA | | Authors: | Rose, R.B, Bayle, J.H, Endrizzi, J.A, Cronk, J.D, Crabtree, G.R, Alber, T. | | Deposit date: | 2000-07-06 | | Release date: | 2000-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of dimerization, coactivator recognition and MODY3 mutations in HNF-1alpha.
Nat.Struct.Biol., 7, 2000
|
|
3OIF
 
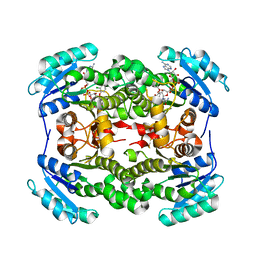 | | Crystal Structure of Enoyl-ACP Reductases I (FabI) from B. subtilis (complex with NAD and TCL) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3EZJ
 
 | | Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody | | Descriptor: | CHLORIDE ION, General secretion pathway protein GspD, NANOBODY NBGSPD_7, ... | | Authors: | Korotkov, K.V, Pardon, E, Steyaert, J, Hol, W.G. | | Deposit date: | 2008-10-22 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody.
Structure, 17, 2009
|
|
3FCB
 
 | |
