3CBX
 
 | |
3CC8
 
 | |
3CCH
 
 | | H-2Db complex with murine gp100 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Badia-Martinez, D, Achour, A. | | Deposit date: | 2008-02-25 | | Release date: | 2009-03-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design of agonistic altered peptides for the robust induction of CTL directed towards H-2Db in complex with the melanoma-associated epitope gp100.
Cancer Res., 69, 2009
|
|
1DKO
 
 | | CRYSTAL STRUCTURE OF TUNGSTATE COMPLEX OF ESCHERICHIA COLI PHYTASE AT PH 6.6 WITH TUNGSTATE BOUND AT THE ACTIVE SITE AND WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE, TUNGSTATE(VI)ION | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
3CCW
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[4-(benzylcarbamoyl)-2-(4-fluorophenyl)-5-(1-methylethyl)-1H-imidazol-1-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
8SBI
 
 | |
6IBM
 
 | | Crystal structure of human alpha-galactosidase A in complex with alpha-galactose configured cyclosulfate ME776 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Rowland, R.J, Wu, L, Davies, G.J. | | Deposit date: | 2018-11-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Alpha-d-Gal-cyclophellitol cyclosulfamidate is a Michaelis complex analog that stabilizes therapeutic lysosomal alpha-galactosidase A in Fabry disease
Chem Sci, 2019
|
|
3CD3
 
 | | Crystal structure of phosphorylated human feline sarcoma viral oncogene homologue (v-FES) in complex with staurosporine and a consensus peptide | | Descriptor: | CHLORIDE ION, Proto-oncogene tyrosine-protein kinase Fes/Fps, STAUROSPORINE, ... | | Authors: | Filippakopoulos, P, Salah, E, Cooper, C, Picaud, S.S, Elkins, J.M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-26 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Coupling of SH2-Kinase Domains Links Fes and Abl Substrate Recognition and Kinase Activation
Cell(Cambridge,Mass.), 134, 2008
|
|
3CD9
 
 | |
6I1A
 
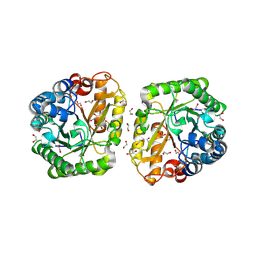 | | Crystal structure of rutinosidase from Aspergillus niger | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, rutinosidase | | Authors: | Pachl, P, Rezacova, P, Kapesova, J. | | Deposit date: | 2018-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Rutinosidase from Aspergillus niger: crystal structure and insight into the enzymatic activity.
Febs J., 287, 2020
|
|
6ICE
 
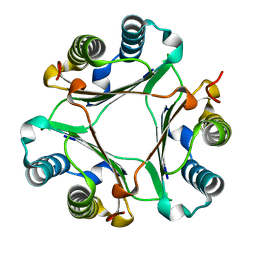 | | Crystal structure of Hamster MIF | | Descriptor: | Macrophage migration inhibitory factor | | Authors: | Sundaram, R, Vasudevan, D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Macrophage migration inhibitory factor of Syrian golden hamster shares structural and functional similarity with human counterpart and promotes pancreatic cancer.
Sci Rep, 9, 2019
|
|
3CDD
 
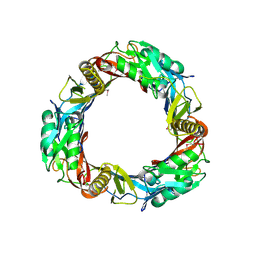 | | Crystal structure of prophage MuSo2, 43 kDa tail protein from Shewanella oneidensis | | Descriptor: | Prophage MuSo2, 43 kDa tail protein | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-26 | | Release date: | 2008-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of prophage MuSo2, 43 kDa tail protein from Shewanella oneidensis.
To be Published
|
|
6I23
 
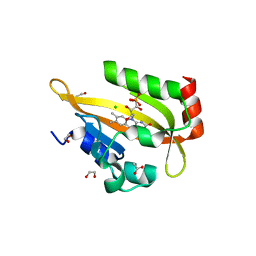 | | Flavin Analogue Sheds Light on Light-Oxygen-Voltage Domain Mechanism | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-5-O-phosphono-D-ribitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Rizkallah, P.J, Kalvaitis, M.E, Allemann, R.K, Mart, R.J, Johnson, L.A. | | Deposit date: | 2018-10-31 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Noncanonical Chromophore Reveals Structural Rearrangements of the Light-Oxygen-Voltage Domain upon Photoactivation.
Biochemistry, 58, 2019
|
|
6DW5
 
 | | SAMHD1 Bound to Gemcitabine-TP in the Catalytic Pocket | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1D9Z
 
 | | CRYSTAL STRUCTURE OF THE DNA REPAIR PROTEIN UVRB IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, EXCINUCLEASE UVRABC COMPONENT UVRB, MAGNESIUM ION, ... | | Authors: | Theis, K, Chen, P.J, Skorvaga, M, Van Houten, B, Kisker, C. | | Deposit date: | 1999-10-30 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair.
EMBO J., 18, 1999
|
|
3CE5
 
 | | A bimolecular parallel-stranded human telomeric quadruplex in complex with a 3,6,9-trisubstituted acridine molecule BRACO19 | | Descriptor: | 9-[4-(n,n-dimethylamino)phenylamino]-3,6-bis(3-pyrrolidinopropionamido) acridine, DNA (5'-D(*DTP*DAP*DGP*DGP*DGP*DTP*DTP*DAP*DGP*DGP*DGP*DT)-3'), POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G.N, Reszka, A.P, Neidle, S. | | Deposit date: | 2008-02-28 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of DNA quadruplex recognition by an acridine drug.
J.Am.Chem.Soc., 130, 2008
|
|
8SX6
 
 | | RNA duplex bound with GMP and AMP monomers | | Descriptor: | ADENOSINE MONOPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
3CE7
 
 | | Crystal structure of toxoplasma specific mitochondrial acyl carrier protein, 59.m03510 | | Descriptor: | Specific mitochondrial acyl carrier protein | | Authors: | Wernimont, A.K, Dong, A, Yang, C, Khuu, C, Lam, A, Brand, V, Kozieradzki, I, Cossar, D, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-28 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of toxoplasma specific mitochondrial acyl carrier protein, 59.m03510.
To be Published
|
|
3CA1
 
 | | Sambucus nigra agglutinin II (SNA-II)- tetragonal crystal form- complexed to galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
3CAL
 
 | |
3CBL
 
 | | Crystal structure of human feline sarcoma viral oncogene homologue (v-FES) in complex with staurosporine and a consensus peptide | | Descriptor: | Proto-oncogene tyrosine-protein kinase Fes/Fps, STAUROSPORINE, Synthetic peptide | | Authors: | Filippakopoulos, P, Salah, E, Cooper, C, Picaud, S.S, Elkins, J.M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-22 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Coupling of SH2-Kinase Domains Links Fes and Abl Substrate Recognition and Kinase Activation
Cell(Cambridge,Mass.), 134, 2008
|
|
6IFU
 
 | | Cryo-EM structure of type III-A Csm-CTR2-dsDNA complex | | Descriptor: | CTR2, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
7S4E
 
 | |
3CBZ
 
 | |
3CCB
 
 | | Crystal Structure of Human DPP4 in complex with a benzimidazole derivative | | Descriptor: | 1-biphenyl-2-ylmethanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wallace, M.B, Skene, R.J. | | Deposit date: | 2008-02-25 | | Release date: | 2008-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-based design and synthesis of benzimidazole derivatives as dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
