8PFE
 
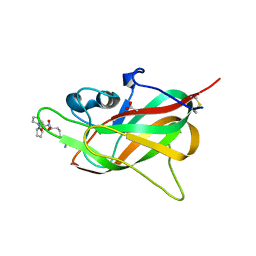 | |
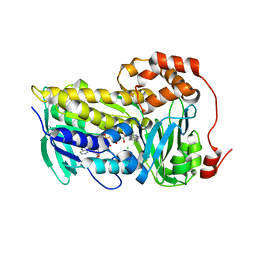
6N0Y
 
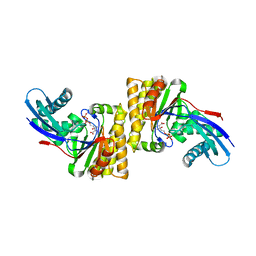 | |
8T3P
 
 | |
7YPP
 
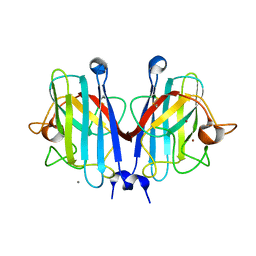 | | Structural basis of a superoxide dismutase from a tardigrade, Ramazzottius varieornatus strain YOKOZUNA-1. | | Descriptor: | CALCIUM ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Sim, K.-S, Fukuda, Y, Inoue, T. | | Deposit date: | 2022-08-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a superoxide dismutase from a tardigrade: Ramazzottius varieornatus strain YOKOZUNA-1.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
7PPX
 
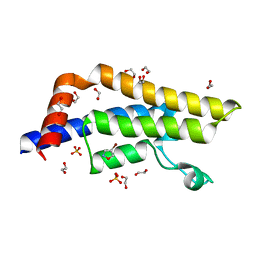 | | ATAD2 in complex with FragLite3 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-1,2-oxazole, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Wood, D.J, Ng, Y.M, Heath, R, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2021-09-15 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
5UQ0
 
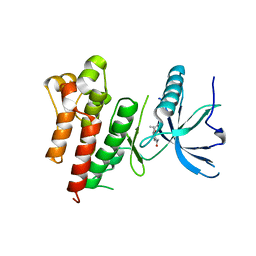 | |
7YW2
 
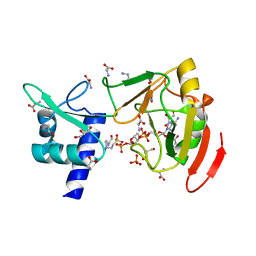 | | Crystal structure of tRNA 2'-phosphotransferase from Mus musculus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, GLYCINE, ... | | Authors: | Yang, X.Y, Liu, X.H. | | Deposit date: | 2022-08-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical insights into the molecular mechanism of TRPT1 for nucleic acid ADP-ribosylation.
Nucleic Acids Res., 51, 2023
|
|
8SX1
 
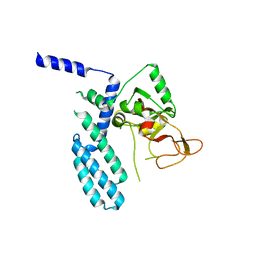 | | PARP4 catalytic domain | | Descriptor: | Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Frigon, L, Pascal, J.M. | | Deposit date: | 2023-05-19 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
8SX2
 
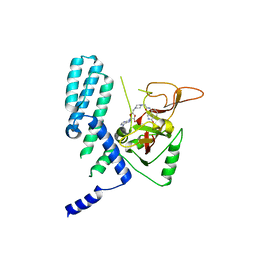 | | PARP4 catalytic domain bound to EB47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Frigon, L, Pascal, J.M. | | Deposit date: | 2023-05-19 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
5HVQ
 
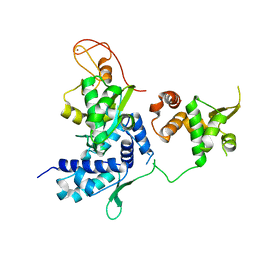 | | Alternative model of the MAGE-G1 NSE-1 complex | | Descriptor: | Melanoma-associated antigen G1, Non-structural maintenance of chromosomes element 1 homolog, ZINC ION | | Authors: | Newman, J.A, Cooper, C.D.O, Roos, A.K, Aitkenhead, H, Oppermann, U.C.T, Cho, H.J, Osman, R, Gileadi, O. | | Deposit date: | 2016-01-28 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Structures of Two Melanoma-Associated Antigens Suggest Allosteric Regulation of Effector Binding.
Plos One, 11, 2016
|
|
8PUN
 
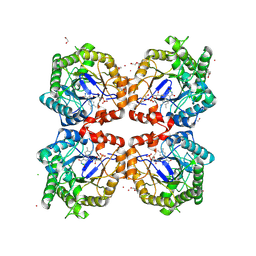 | |
7ZC9
 
 | | Human Pikachurin/EGFLAM C-terminal Laminin-G domain (LG3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pikachurin, SULFATE ION | | Authors: | Pantalone, S, Forneris, F. | | Deposit date: | 2022-03-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179.
Sci.Signal., 16, 2023
|
|
7ZCB
 
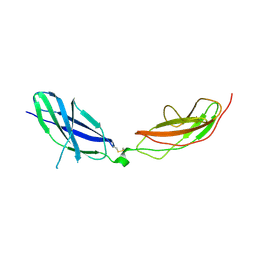 | | Human Pikachurin/EGFLAM N-terminal Fibronectin-III (1-2) domains | | Descriptor: | CHLORIDE ION, Pikachurin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Pantalone, S, Savino, S, Viti, L.V, Forneris, F. | | Deposit date: | 2022-03-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179.
Sci.Signal., 16, 2023
|
|
7PSF
 
 | |
8VEX
 
 | |
7YKC
 
 | |
8PPZ
 
 | | Co-crystal structure of FKBP12, compound 7 and the FRB fragment of mTOR | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-[(~{E})-2-(2-chlorophenyl)ethenyl]-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Meyners, C, Deutscher, R.C.E, Hausch, F. | | Deposit date: | 2023-07-10 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Co-crystal structure of FKBP12, compound 7 and the FRB fragment of mTOR
Chemrxiv, 2023
|
|
7PRK
 
 | |
7NFG
 
 | | A hexameric barrel state of a de novo coiled-coil assembly: CC-Type2-(LaId)4-L14A. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CC-Type2-(LaId)4-L14A | | Authors: | Rhys, G.G, Dawson, W.M, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2021-02-07 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Differential sensing with arrays of de novo designed peptide assemblies.
Nat Commun, 14, 2023
|
|
7NFH
 
 | | A heptameric barrel state of a de novo coiled-coil assembly: CC-Type2-(MaId)4. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CC-Type2-(MaId)4 | | Authors: | Rhys, G.G, Dawson, W.M, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2021-02-07 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Differential sensing with arrays of de novo designed peptide assemblies.
Nat Commun, 14, 2023
|
|
8V9Q
 
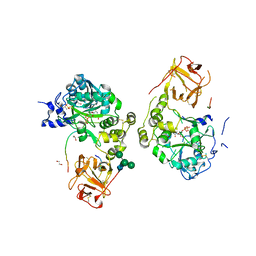 | | Crystal structure of mGalNAc-T1 in complex with the mucin glycopeptide Muc5AC-13, Mn2+, and UDP. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Samara, N.L, Collette, A.M. | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | An unusual dual sugar-binding lectin domain controls the substrate specificity of a mucin-type O-glycosyltransferase.
Sci Adv, 10, 2024
|
|
6N4Y
 
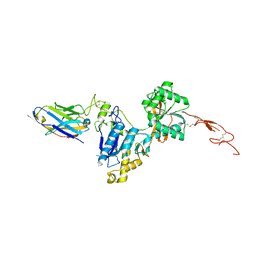 | | Metabotropic Glutamate Receptor 5 Extracellular Domain with Nb43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Chu, M, Weis, W.I, Mathiesen, J.M, Skiniotis, G, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.262 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
8SWY
 
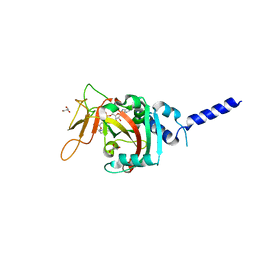 | | PARP4 ART domain bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Frigon, L, Pascal, J.M. | | Deposit date: | 2023-05-19 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
8VCN
 
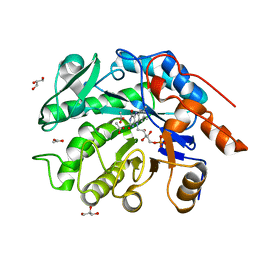 | | GluER mutant - W66F F269Y Q293T F68Y T36E P263L | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Jeffrey, P.D, Sorigue, D.R, Liu, Y, Hyster, T.K. | | Deposit date: | 2023-12-14 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Asymmetric Synthesis of alpha-Chloroamides via Photoenzymatic Hydroalkylation of Olefins.
J.Am.Chem.Soc., 146, 2024
|
|
8VRS
 
 | | Mucin 16 peptide fused to MBP in complex with 4H11-scFv antibody | | Descriptor: | 4H11 scFv chain, Maltose/maltodextrin-binding periplasmic protein,Mucin-16 | | Authors: | Lee, K, Perry, K, Yeku, O.O, Spriggs, D.R. | | Deposit date: | 2024-01-22 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural basis for antibody recognition of the proximal MUC16 ectodomain.
J Ovarian Res, 17, 2024
|
|
