3BTZ
 
 | | Crystal structure of human ABH2 cross-linked to dsDNA | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*AP*GP*GP*TP*GP*AP*(2YR)P*AP*AP*TP*GP*CP*G)-3'), DNA (5'-D(*DTP*DCP*DGP*DCP*DAP*DTP*DTP*DAP*DTP*DCP*DAP*DCP*DC)-3') | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
5DI2
 
 | | Two divalent metal ions and conformational changes play roles in the hammerhead ribozyme cleavage reaction-WT ribozyme in Mn2+ at high pH | | Descriptor: | 5'-R(*GP*GP*GP*CP*GP*U)-D(P*C)-R(P*UP*GP*GP*GP*CP*AP*GP*UP*AP*CP*CP*CP*A)-3', MANGANESE (II) ION, RNA (48-MER) | | Authors: | Mir, A, Chen, J, Neau, D, Golden, B.L. | | Deposit date: | 2015-08-31 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Two Divalent Metal Ions and Conformational Changes Play Roles in the Hammerhead Ribozyme Cleavage Reaction.
Biochemistry, 54, 2015
|
|
3BUC
 
 | | X-ray structure of human ABH2 bound to dsDNA with Mn(II) and 2KG | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*DCP*DTP*DGP*DTP*DAP*DTP*(MA7)P*DAP*DCP*DTP*DGP*DCP*DG)-3'), ... | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2008-01-02 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
1XXT
 
 | | The T-to-T High Transitions in Human Hemoglobin: wild-type deoxy Hb A (low salt, one test set) | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Rogers, P.H, Arnone, A. | | Deposit date: | 2004-11-08 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystallographic evidence for a new ensemble of ligand-induced allosteric transitions in hemoglobin: the T-to-T(high) quaternary transitions.
Biochemistry, 44, 2005
|
|
2PQG
 
 | | Crystal structure of inactive ribosome inactivating protein from maize (b-32) | | Descriptor: | Ribosome-inactivating protein 3 | | Authors: | Mak, A.N.S, Wong, Y.T, Young, J.A, Cha, S.S, Sze, K.H, Au, S.W.N, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-05-02 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure-function study of maize ribosome-inactivating protein: implications for the internal inactivation region and the sole glutamate in the active site.
Nucleic Acids Res., 35, 2007
|
|
6R47
 
 | |
3BU0
 
 | | crystal structure of human ABH2 cross-linked to dsDNA with cofactors | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*CP*TP*GP*TP*AP*TP*(2YR)P*AP*TP*TP*GP*CP*G)-3'), ... | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
3BTX
 
 | | X-ray structure of human ABH2 bound to dsDNA through active site cross-linking | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*CP*TP*GP*TP*AP*TP*(2YR)P*AP*TP*TP*GP*CP*G)-3'), DNA (5'-D(*DTP*DCP*DGP*DCP*DAP*DAP*DTP*DAP*DAP*DTP*DAP*DCP*DA)-3'), ... | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
2PQI
 
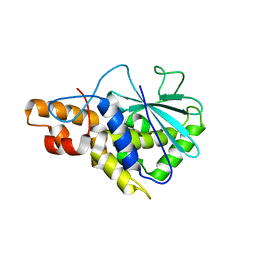 | | Crystal structure of active ribosome inactivating protein from maize (b-32) | | Descriptor: | Ribosome-inactivating protein 3 | | Authors: | Mak, A.N.S, Wong, Y.T, Young, J.A, Cha, S.S, Sze, K.H, Au, S.W.N, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-05-02 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function study of maize ribosome-inactivating protein: implications for the internal inactivation region and the sole glutamate in the active site.
Nucleic Acids Res., 35, 2007
|
|
3L7V
 
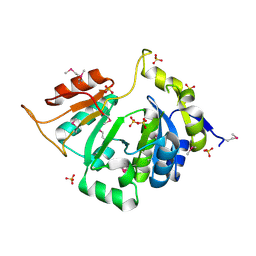 | |
3DCM
 
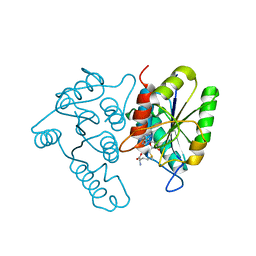 | | Crystal structure of the Thermotoga maritima SPOUT family RNA-methyltransferase protein Tm1570 in complex with S-adenosyl-L-methionine | | Descriptor: | S-ADENOSYLMETHIONINE, Uncharacterized protein TM_1570 | | Authors: | Kim, D.J, Kim, H.S, Lee, S.J, Suh, S.W. | | Deposit date: | 2008-06-04 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Thermotoga maritima SPOUT superfamily RNA methyltransferase Tm1570 in complex with S-adenosyl-L-methionine
Proteins, 74, 2009
|
|
4XEM
 
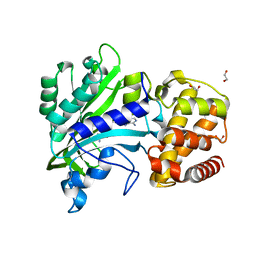 | |
4XEO
 
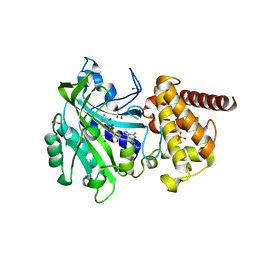 | |
7ZJ4
 
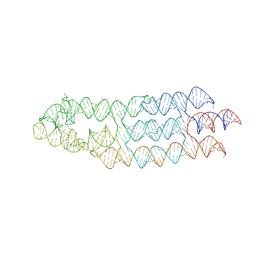 | | Ligand bound state of a brocolli-pepper aptamer FRET tile | | Descriptor: | 4-(3,5-difluoro-4-hydroxybenzyl)-1,2-dimethyl-1H-imidazol-5-ol, 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]thieno[3,2-b]thiophen-2-yl]ethenyl]benzenecarbonitrile, POTASSIUM ION, ... | | Authors: | McRae, E.K.S, Vallina, N.S, Hansen, B.K, Boussebayle, A, Andersen, E.S. | | Deposit date: | 2022-04-08 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structure determination of Pepper-Broccoli FRET pair by RNA origami scaffolding
To Be Published
|
|
6YI9
 
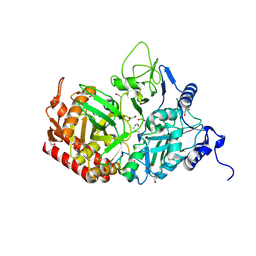 | | Crystal structure of the rat cytosolic PCK1, acetylated on Lys244 | | Descriptor: | 1,2-ETHANEDIOL, Phosphoenolpyruvate carboxykinase, cytosolic [GTP] | | Authors: | Latorre-Muro, P, Baeza, J, Hurtado-Guerrero, R, Hicks, T, Delso, I, Hernandez-Ruiz, C, Velazquez-Campoy, A, Lawton, A.J, Angulo, J, Denu, J.M, Carrodeguas, J.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Self-acetylation at the active site of phosphoenolpyruvate carboxykinase (PCK1) controls enzyme activity.
J.Biol.Chem., 296, 2021
|
|
7L4X
 
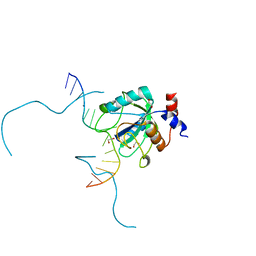 | |
7L4Y
 
 | |
8E2R
 
 | | Crystal structure of TadAC-1.14 | | Descriptor: | GLYCEROL, ZINC ION, tRNA-specific adenosine deaminase 1.14 | | Authors: | Feliciano, P.R, Lee, S.J, Ciaramella, G. | | Deposit date: | 2022-08-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Improved cytosine base editors generated from TadA variants.
Nat.Biotechnol., 41, 2023
|
|
8E2P
 
 | |
8E2S
 
 | |
8E2Q
 
 | | Crystal structure of TadAC-1.17 in a complex with ssDNA | | Descriptor: | DNA (5'-D(P*GP*CP*GP*GP*CP*TP*(D8A)P*CP*GP*GP*A)-3'), GLYCEROL, ZINC ION, ... | | Authors: | Feliciano, P.R, Lee, S.J, Ciaramella, G. | | Deposit date: | 2022-08-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Improved cytosine base editors generated from TadA variants.
Nat.Biotechnol., 41, 2023
|
|
1Q5K
 
 | | crystal structure of Glycogen synthase kinase 3 in complexed with inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-(4-METHOXYBENZYL)-N'-(5-NITRO-1,3-THIAZOL-2-YL)UREA | | Authors: | Bhat, R, Xue, Y, Berg, S, Hellberg, S, Ormo, M, Nilsson, Y, Radesater, A.C, Jerning, E, Markgren, P.O, Borgegard, T, Nylof, M, Gimenez-Cassina, A, Hernandez, F, Lucas, J.J, Diaz-Mido, J, Avila, J. | | Deposit date: | 2003-08-08 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-A014418.
J.Biol.Chem., 278, 2003
|
|
3G8S
 
 | | Crystal structure of the pre-cleaved Bacillus anthracis glmS ribozyme | | Descriptor: | GLMS RIBOZYME, MAGNESIUM ION, RNA (5'-R(*AP*(A2M)P*GP*CP*GP*CP*CP*AP*GP*AP*AP*CP*U)-3'), ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3G96
 
 | | Crystal structure of the Bacillus anthracis glmS ribozyme bound to MaN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
2GIY
 
 | |
