7Y6L
 
 | |
7Y6N
 
 | |
7YBJ
 
 | | SARS-CoV-2 Mu variant spike(close state) | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBI
 
 | | SARS-CoV-2 Mu variant spike (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7Y8J
 
 | | 3D1 in complex with 6-mer HR1 peptide from SARS-CoV-2 | | Descriptor: | Spike protein S2', heavy chain of 3D1, light chain of 3D1 | | Authors: | Lei, Y. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Cross-reactive epitopes between HIV and Coronavirus revealed by 3D1
To Be Published
|
|
7Y6K
 
 | |
7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
7Y72
 
 | | SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 (focused refinement on Fab-RBD interface) | | Descriptor: | Fab E7 heavy chain, Fab E7 light chain, Spike glycoprotein | | Authors: | Chia, W.N, Tan, C.W, Tan, A.W.K, Young, B, Starr, T.N, Lopez, E, Fibriansah, G, Barr, J, Cheng, S, Yeoh, A.Y.Y, Yap, W.C, Lim, B.L, Ng, T.S, Sia, W.R, Zhu, F, Chen, S, Zhang, J, Greaney, A.J, Chen, M, Au, G.G, Paradkar, P, Peiris, M, Chung, A.W, Bloom, J.D, Lye, D, Lok, S.M, Wang, L.F. | | Deposit date: | 2022-06-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Potent pan huACE2-dependent sarbecovirus neutralizing monoclonal antibodies isolated from a BNT162b2-vaccinated SARS survivor.
Sci Adv, 9, 2023
|
|
7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
7YDY
 
 | | SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YBL
 
 | | SARS-CoV-2 B.1.620 variant spike (close state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7Y71
 
 | | SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab E7 heavy chain, ... | | Authors: | Chia, W.N, Tan, C.W, Tan, A.W.K, Young, B, Starr, T.N, Lopez, E, Fibriansah, G, Barr, J, Cheng, S, Yeoh, A.Y.Y, Yap, W.C, Lim, B.L, Ng, T.S, Sia, W.R, Zhu, F, Chen, S, Zhang, J, Greaney, A.J, Chen, M, Au, G.G, Paradkar, P, Peiris, M, Chung, A.W, Bloom, J.D, Lye, D, Lok, S.M, Wang, L.F. | | Deposit date: | 2022-06-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Potent pan huACE2-dependent sarbecovirus neutralizing monoclonal antibodies isolated from a BNT162b2-vaccinated SARS survivor.
Sci Adv, 9, 2023
|
|
7YBK
 
 | | SARS-CoV-2 B.1.620 variant spike (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7Y0O
 
 | |
7YBH
 
 | | SARS-CoV-2 lambda variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBM
 
 | | SARS-CoV-2 C.1.2 variant spike (Close state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7XY4
 
 | |
7XY3
 
 | |
7Y0N
 
 | | SARS-CoV-2 WT Spike in complex with R15 Fab and P14 Nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R15-F7, ... | | Authors: | Wang, L. | | Deposit date: | 2022-06-05 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-CoV-2 WT Spike in complex with R15 Fab and P14 Nanobody
To Be Published
|
|
7WD1
 
 | | Crystal structure of R14 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, R14, Spike protein S1, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
7WD2
 
 | | Crystal structure of S43 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
7WGZ
 
 | | SARS-CoV-2 spike glycoprotein trimer in open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
7WGX
 
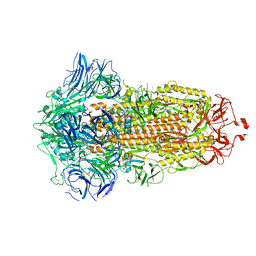 | | SARS-CoV-2 spike glycoprotein trimer in closed state after treatment with Cathepsin L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
7WGY
 
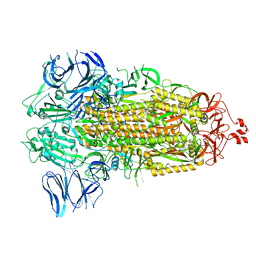 | | SARS-CoV-2 spike glycoprotein trimer in Intermediate state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
7WGV
 
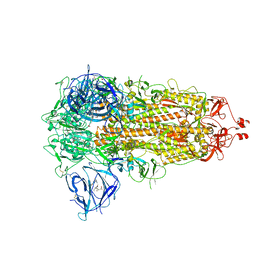 | | SARS-CoV-2 spike glycoprotein trimer in closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
