7MNO
 
 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 1-752) I656V mutant in complex with Fab fragment | | Descriptor: | Antibody Fab14 Heavy Chain, Antibody Fab14 Light Chain, E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (6.73 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7YEN
 
 | |
8K7O
 
 | | De novo design protein -T03 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo design protein -T03
To Be Published
|
|
6MKQ
 
 | | Carbapenemase VCC-1 bound to avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Mark, B.L, Vadlamani, G. | | Deposit date: | 2018-09-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for the Potent Inhibition of the Emerging Carbapenemase VCC-1 by Avibactam.
Antimicrob. Agents Chemother., 63, 2019
|
|
7P2Z
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with cyclosulfamidate 4 | | Descriptor: | (3~{a}~{R},4~{S},5~{S},6~{S},7~{R},7~{a}~{S})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydro-3~{H}-benzo[d][1,2,3]oxathiazole-4,5,6-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Kok, K, Overkleeft, H, Artola, M, Sulzenbacher, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,6- epi-Cyclophellitol Cyclosulfamidate Is a Bona Fide Lysosomal alpha-Glucosidase Stabilizer for the Treatment of Pompe Disease.
J.Am.Chem.Soc., 144, 2022
|
|
6ML6
 
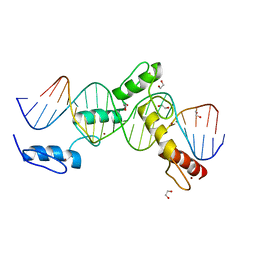 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpA 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*(5CM)P*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
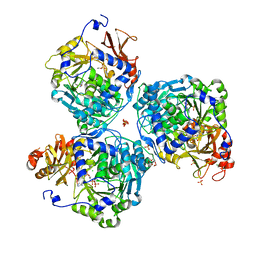
8V4P
 
 | |
7P0U
 
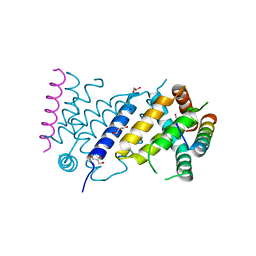 | | ORF virus encoded Bcl-2 homolog ORFV125 in complex with Puma BH3 peptide | | Descriptor: | 1,2-ETHANEDIOL, Activator of apoptosis harakiri, Apoptosis inhibitor, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99374259 Å) | | Cite: | Structural Investigation of Orf Virus Bcl-2 Homolog ORFV125 Interactions with BH3-Motifs from BH3-Only Proteins Puma and Hrk.
Viruses, 13, 2021
|
|
7YE3
 
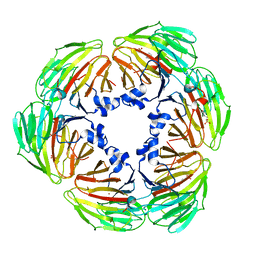 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
5UDD
 
 | | Crystal Structure of RSV F B9320 Bound to MEDI8897 | | Descriptor: | Fusion glycoprotein F0, MEDI8897 Fab Heavy Chain, MEDI8897 Fab Light Chain, ... | | Authors: | McLellan, J.S. | | Deposit date: | 2016-12-26 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants.
Sci Transl Med, 9, 2017
|
|
8K48
 
 | | LTD of arabidopsis thaliana | | Descriptor: | Protein LHCP TRANSLOCATION DEFECT | | Authors: | Wang, C, Xu, X.M. | | Deposit date: | 2023-07-17 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Arabidopsis thaliana LTD
To Be Published
|
|
8USJ
 
 | |
5H2W
 
 | |
7V6D
 
 | | Structure of lipase B from Lasiodiplodia theobromae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lipase B | | Authors: | Xue, B, Zhang, H.F, Nguyen, G.K.T, Yew, W.S. | | Deposit date: | 2021-08-20 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Lipase from Lasiodiplodia theobromae Efficiently Hydrolyses C8-C10 Methyl Esters for the Preparation of Medium-Chain Triglycerides' Precursors.
Int J Mol Sci, 22, 2021
|
|
8UZ8
 
 | |
8SA9
 
 | |
8SBY
 
 | | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (NAD and sulfate bound, hexagonal form) | | Descriptor: | 2,3-dihydroxybenzoate-2,3-dehydrogenase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2023-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (NAD and sulfate bound, hexagonal form)
To be published
|
|
7P0S
 
 | |
7MNN
 
 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 1-752) T653I mutant in complex with Fab fragment | | Descriptor: | Antibody Fab14 Heavy Chain, Antibody Fab14 Light Chain, E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (6.7 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
8USH
 
 | |
7VKO
 
 | | Crystal structure of TrkA kinase with repotrectinib | | Descriptor: | Repotrectinib, SULFATE ION, Tyrosine-protein kinase receptor | | Authors: | Murray, B.W, Rogers, E, Zhai, D, Deng, W, Chen, X, Sprengeler, P.A, Zhang, X, Graber, A, Reich, S.H, Stopatschinskaja, S, Solomon, B, Besse, B, Drilon, A. | | Deposit date: | 2021-09-30 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Characteristics of Repotrectinib That Enable Potent Inhibition of TRK Fusion Proteins and Resistant Mutations.
Mol.Cancer Ther., 20, 2021
|
|
7VKN
 
 | | Crystal structure of TrkA (G595R) kinase with repotrectinib | | Descriptor: | Repotrectinib, SULFATE ION, Tyrosine-protein kinase receptor | | Authors: | Murray, B.W, Rogers, E, Zhai, D, Deng, W, Chen, X, Sprengeler, P.A, Zhang, X, Graber, A, Reich, S.H, Stopatschinskaja, S, Solomon, B, Besse, B, Drilon, A. | | Deposit date: | 2021-09-30 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Characteristics of Repotrectinib That Enable Potent Inhibition of TRK Fusion Proteins and Resistant Mutations.
Mol.Cancer Ther., 20, 2021
|
|
8V15
 
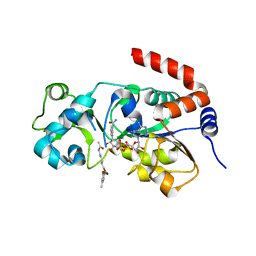 | | Human SIRT3 bound to p53-AMC peptide, Carba-NAD, and Honokiol | | Descriptor: | (1P)-3',5-di(prop-2-en-1-yl)[1,1'-biphenyl]-2,4'-diol, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLN-PRO-LYS-FDL, ... | | Authors: | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | Deposit date: | 2023-11-19 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computationally Driven Discovery and Characterization of SIRT3 Activating Compounds that Fully Recover Catalytic Activity under NAD+ Depletion
biorxiv, 2023
|
|
8KCN
 
 | |
6MP5
 
 | |
