3D1I
 
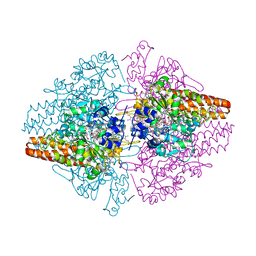 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with nitrite | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-05-06 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens
J.Mol.Biol., 389, 2009
|
|
5OBH
 
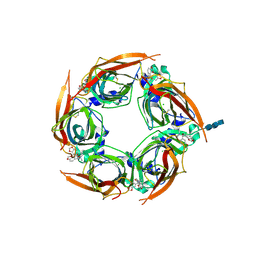 | | Crystal structure of glycine binding protein in complex with bicuculline | | Descriptor: | (5S)-6,6-dimethyl-5-[(6R)-8-oxo-6,8-dihydrofuro[3,4-e][1,3]benzodioxol-6-yl]-5,6,7,8-tetrahydro[1,3]dioxolo[4,5-g]isoquinolin-6-ium, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dawson, A, Hunter, W.N, Jones, M. | | Deposit date: | 2017-06-27 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Structural Rationale for N-Methylbicuculline Acting as a Promiscuous Competitive Antagonist of Inhibitory Pentameric Ligand-Gated Ion Channels.
Chembiochem, 21, 2020
|
|
3C7F
 
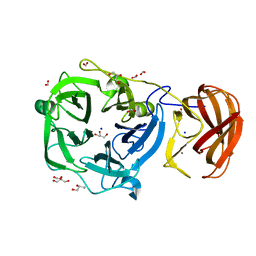 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from bacillus subtilis in complex with xylotriose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3UZR
 
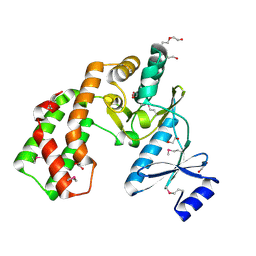 | | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, apo form | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside phosphotransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Minasov, G, Singer, A.U, Tan, K, Nocek, B, Evdokimova, E, Egorova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-07 | | Release date: | 2011-12-21 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, apo form
TO BE PUBLISHED
|
|
3CMT
 
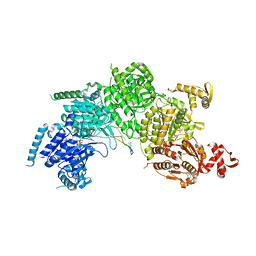 | | Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*DTP*DTP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DCP*DTP*DTP*DTP*DT)-3'), DNA (5'-D(P*DGP*DGP*DTP*DGP*DGP*DG)-3'), ... | | Authors: | Chen, Z, Yang, H, Pavletich, N.P. | | Deposit date: | 2008-03-24 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures.
Nature, 453, 2008
|
|
3CSB
 
 | |
3C64
 
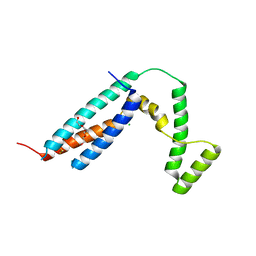 | | The MC179 portion of the Cysteine-rich Interdomain Region (CIDR) of a Plasmodium falciparum Erythrocyte Membrane Protein-1 (PfEMP1) | | Descriptor: | CHLORIDE ION, PfEMP1 variant 2 of strain MC, TETRAETHYLENE GLYCOL | | Authors: | Klein, M.M, Gittis, A.G, Su, H.P, Makobongo, M.O, Moore, J.M, Singh, S, Miller, L.H, Garboczi, D.N. | | Deposit date: | 2008-02-02 | | Release date: | 2008-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Cysteine-Rich Interdomain Region from the Highly Variable Plasmodium falciparum Erythrocyte Membrane Protein-1 Exhibits a Conserved Structure.
Plos Pathog., 4, 2008
|
|
1KPM
 
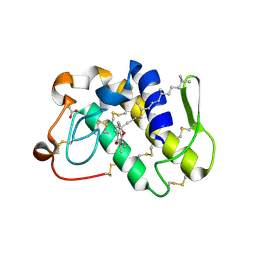 | | First Structural Evidence of a Specific Inhibition of Phospholipase A2 by Vitamin E and its Implications in Inflammation: Crystal Structure of the Complex Formed between Phospholipase A2 and Vitamin E at 1.8 A Resolution. | | Descriptor: | ACETIC ACID, Phospholipase A2, VITAMIN E | | Authors: | Chandra, V, Jasti, J, Kaur, P, Betzel, C, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-01-01 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | First Structural Evidence of a Specific Inhibition of Phospholipase A2 by alpha-Tocopherol (Vitamin E) and its
Implications in Inflammation: Crystal Structure of the Complex Formed Between Phospholipase A2 and
alpha-Tocopherol at 1.8 A Resolution
J.Mol.Biol., 320, 2002
|
|
4GP1
 
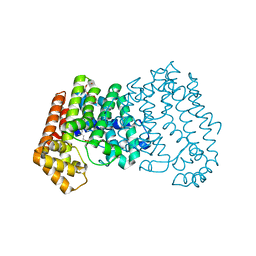 | | Crystal structure of ISOPRENOID SYNTHASE A3MSH1 (TARGET EFI-501992) from pyrobaculum calidifontis complexed with DMAPP | | Descriptor: | CHLORIDE ION, DIMETHYLALLYL DIPHOSPHATE, Polyprenyl synthetase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Isoprenoid Synthase from Pyrobaculum Calidifontis
To be Published
|
|
2P3G
 
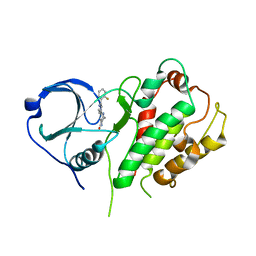 | | Crystal structure of a pyrrolopyridine inhibitor bound to MAPKAP Kinase-2 | | Descriptor: | 2-[2-(2-FLUOROPHENYL)PYRIDIN-4-YL]-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP kinase-activated protein kinase 2 | | Authors: | Kurumbail, R.G, Caspers, N. | | Deposit date: | 2007-03-08 | | Release date: | 2007-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Pyrrolopyridine Inhibitors of Mitogen-Activated Protein Kinase-Activated Protein Kinase 2 (MK-2).
J.Med.Chem., 50, 2007
|
|
3UPC
 
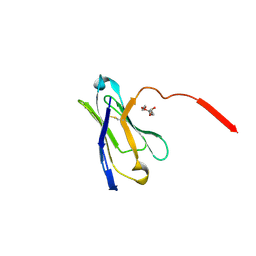 | | A general strategy for the generation of human antibody variable domains with increased aggregation resistance | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | Dudgeon, K, Rouet, R, Kokmeijer, I, Langley, D.B, Christ, D. | | Deposit date: | 2011-11-17 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | General strategy for the generation of human antibody variable domains with increased aggregation resistance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4H41
 
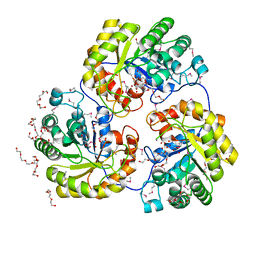 | |
2OU9
 
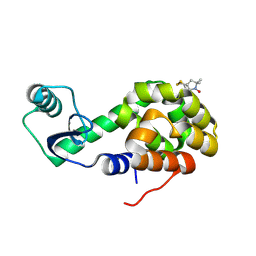 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R1/R119A | | Descriptor: | Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
3DUS
 
 | | Crystal structure of SAG506-01, orthorhombic, twinned, crystal 1 | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, Ig-like protein, MAGNESIUM ION, ... | | Authors: | Brooks, C.L, Blackler, R.J, Gerstenbruch, S, Kosma, P, Muller-Loennies, S, Brade, H, Evans, S.V. | | Deposit date: | 2008-07-17 | | Release date: | 2008-12-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pseudo-symmetry and twinning in crystals of homologous antibody Fv fragments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3DZZ
 
 | |
2Q7H
 
 | | Pyrrolysyl-tRNA synthetase bound to adenylated pyrrolysine and pyrophosphate | | Descriptor: | (2R)-2-AMINO-6-({[(2S,3R)-3-METHYLPYRROLIDIN-2-YL]CARBONYL}AMINO)HEXANOYL [(2S,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL HYDROGEN (R)-PHOSPHATE, 1,2-ETHANEDIOL, PYROPHOSPHATE 2-, ... | | Authors: | Kavran, J.M, Steitz, T.A. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2Q88
 
 | | Crystal structure of EhuB in complex with ectoine | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CADMIUM ION, Putative ABC transporter amino acid-binding protein | | Authors: | Hanekop, N, Hoeing, M, Sohn-Bosser, L, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2007-06-09 | | Release date: | 2008-01-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ligand-binding protein EhuB from Sinorhizobium meliloti reveals substrate recognition of the compatible solutes ectoine and hydroxyectoine.
J.Mol.Biol., 374, 2007
|
|
3ZUA
 
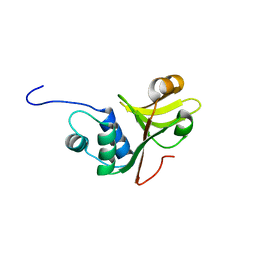 | | A C39-like domain | | Descriptor: | ALPHA-HEMOLYSIN TRANSLOCATION ATP-BINDING PROTEIN HLYB | | Authors: | Lecher, J, Schwarz, C.K.W, Stoldt, M, Smits, S.S.H, Willbold, D, Schmitt, L. | | Deposit date: | 2011-07-18 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An Rtx Transporter Tethers its Unfolded Substrate During Secretion Via a Unique N-Terminal Domain.
Structure, 20, 2012
|
|
5OSS
 
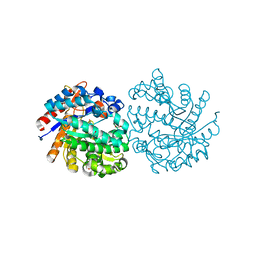 | | Beta-glucosidase from Thermotoga maritima in complex with Gluco-1H-imidazole | | Descriptor: | (4~{S},5~{S},6~{R},7~{R})-7-(hydroxymethyl)-4,5,6,7-tetrahydro-1~{H}-benzimidazole-4,5,6-triol, 1,2-ETHANEDIOL, Beta-glucosidase A, ... | | Authors: | Offen, W.A, Schroeder, S.P, Davies, G.J, Overkleeft, H.S. | | Deposit date: | 2017-08-18 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gluco-1 H-imidazole: A New Class of Azole-Type beta-Glucosidase Inhibitor.
J. Am. Chem. Soc., 140, 2018
|
|
2Q2O
 
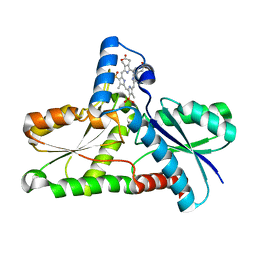 | | Crystal structure of H183C Bacillus subtilis ferrochelatase in complex with deuteroporphyrin IX 2,4-disulfonic acid dihydrochloride | | Descriptor: | Ferrochelatase, MAGNESIUM ION, PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-29 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
5P9F
 
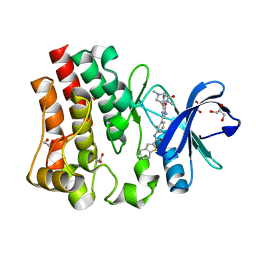 | | BTK IN COMPLEX WITH GDC-0834 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-{3-[6-({4-[(2R)-1,4-dimethyl-3-oxopiperazin-2-yl]phenyl}amino)-4-methyl-5-oxo-4,5-dihydropyrazin-2-yl]-2-methylphenyl }-4,5,6,7-tetrahydro-1-benzothiophene-2-carboxamide, ... | | Authors: | Gardberg, A.S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
Mol. Pharmacol., 91, 2017
|
|
3DUR
 
 | | Crystal structure of SAG173-04 | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, Ig-like protein, MAGNESIUM ION, ... | | Authors: | Brooks, C.L, Blackler, R.J, Gerstenbruch, S, Kosma, P, Muller-Loennies, S, Brade, H, Evans, S.V. | | Deposit date: | 2008-07-17 | | Release date: | 2008-12-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pseudo-symmetry and twinning in crystals of homologous antibody Fv fragments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4IOX
 
 | | The structure of the herpes simplex virus DNA-packaging motor pUL15 C-terminal nuclease domain provides insights into cleavage of concatemeric viral genome precursors | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Selvarajan Sigamani, S, Zhao, H, Kamau, Y, Tang, L. | | Deposit date: | 2013-01-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.458 Å) | | Cite: | The Structure of the Herpes Simplex Virus DNA-Packaging Terminase pUL15 Nuclease Domain Suggests an Evolutionary Lineage among Eukaryotic and Prokaryotic Viruses.
J.Virol., 87, 2013
|
|
2PMO
 
 | | Crystal structure of PfPK7 in complex with hymenialdisine | | Descriptor: | 4-(5-AMINO-4-OXO-4H-PYRAZOL-3-YL)-2-BROMO-4,5,6,7-TETRAHYDRO-3AH-PYRROLO[2,3-C]AZEPIN-8-ONE, Ser/Thr protein kinase | | Authors: | Merckx, A, Echalier, A, Noble, M, Endicott, J. | | Deposit date: | 2007-04-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of P. falciparum protein kinase 7 identify an activation motif and leads for inhibitor design.
Structure, 16, 2008
|
|
3ZC6
 
 | | Crystal structure of JAK3 kinase domain in complex with an indazole substituted pyrrolopyrazine inhibitor | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, N-[(2R)-1-(3-cyanoazetidin-1-yl)-1-oxidanylidene-propan-2-yl]-2-(6-fluoranyl-1-methyl-indazol-3-yl)-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, ... | | Authors: | Kuglstatter, A, Jestel, A, Nagel, S, Boettcher, J, Blaesse, M. | | Deposit date: | 2012-11-16 | | Release date: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Strategic Use of Conformational Bias and Structure Based Design to Identify Potent Jak3 Inhibitors with Improved Selectivity Against the Jak Family and the Kinome.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
