7BV1
 
 | | Cryo-EM structure of the apo nsp12-nsp7-nsp8 complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
7BV2
 
 | | The nsp12-nsp7-nsp8 complex bound to the template-primer RNA and triphosphate form of Remdesivir(RTP) | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
7BF4
 
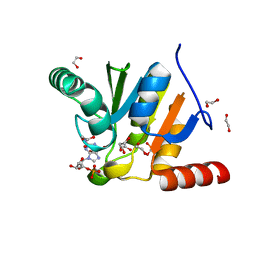 | | Crystal structure of SARS-CoV-2 macrodomain in complex with GMP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, NSP3 macrodomain | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
7BWJ
 
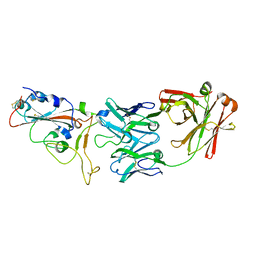 | | crystal structure of SARS-CoV-2 antibody with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Wang, X, Ge, J. | | Deposit date: | 2020-04-14 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Human neutralizing antibodies elicited by SARS-CoV-2 infection.
Nature, 584, 2020
|
|
7BYR
 
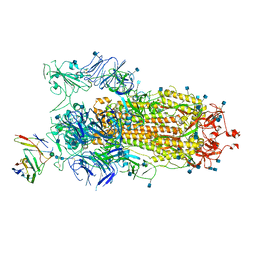 | | BD23-Fab in complex with the S ectodomain trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab23-Fab-Heavy Chain, ... | | Authors: | Zhu, Q, Wang, G, Xiao, J. | | Deposit date: | 2020-04-24 | | Release date: | 2020-06-10 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients' B Cells.
Cell, 182, 2020
|
|
7BW4
 
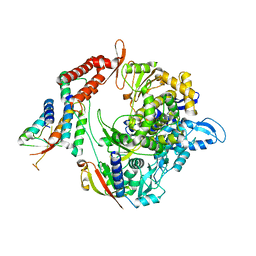 | | Structure of the RNA-dependent RNA polymerase from SARS-CoV-2 | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Peng, Q, Peng, R, Shi, Y. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and Biochemical Characterization of the nsp12-nsp7-nsp8 Core Polymerase Complex from SARS-CoV-2.
Cell Rep, 31, 2020
|
|
7BH9
 
 | | SARS-CoV-2 RBD-62 in complex with ACE2 peptidase domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Elad, N, Dym, O, Zahradnik, J, Schreiber, G. | | Deposit date: | 2021-01-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution.
Nat Microbiol, 6, 2021
|
|
7BE7
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7BGP
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in absence of DTT. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-01-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7BB2
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.6A resolution (spacegroup P2(1)2(1)2(1)) | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
6ZON
 
 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 1 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZLW
 
 | | SARS-CoV-2 Nsp1 bound to the human 40S ribosomal subunit | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-01 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZP1
 
 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7AD1
 
 | | Cryo-EM structure of a prefusion stabilized SARS-CoV-2 Spike (D614N, R682S, R685G, A892P, A942P and V987P)(One up trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein,Spike glycoprotein,Envelope glycoprotein,SARS-CoV-2 S protein | | Authors: | Rutten, L, Renault, L.L.R, Juraszek, J, Langedijk, J.P.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Stabilizing the closed SARS-CoV-2 spike trimer.
Nat Commun, 12, 2021
|
|
6ZN5
 
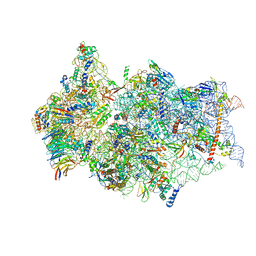 | | SARS-CoV-2 Nsp1 bound to a pre-40S-like ribosome complex - state 2 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZOY
 
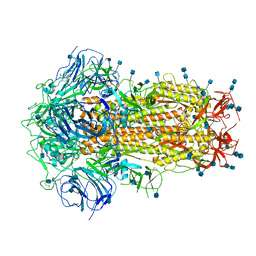 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZP4
 
 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 2 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
7BF5
 
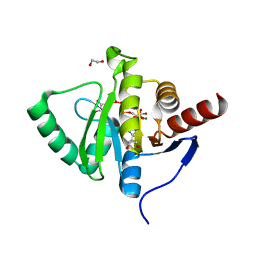 | | Crystal structure of SARS-CoV-2 macrodomain in complex with ADP-ribose-phosphate (ADP-ribose-2'-phosphate, ADPRP) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NSP3 macrodomain, ... | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6ZME
 
 | | SARS-CoV-2 Nsp1 bound to the human CCDC124-80S-eERF1 ribosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZMT
 
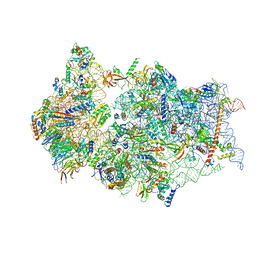 | | SARS-CoV-2 Nsp1 bound to a pre-40S-like ribosome complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZP0
 
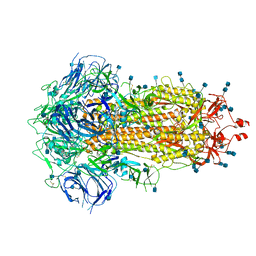 | | Structure of SARS-CoV-2 Spike Protein Trimer (single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZPE
 
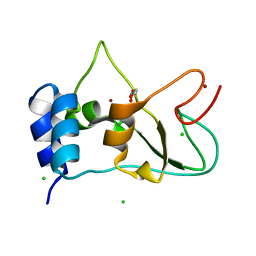 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Replicase polyprotein 1ab, ... | | Authors: | Fisher, S.Z, Kozielski, F. | | Deposit date: | 2020-07-08 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
6ZOX
 
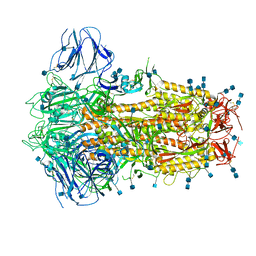 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x2 disulphide-bond mutant, G413C, V987C, single Arg S1/S2 cleavage site) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZP2
 
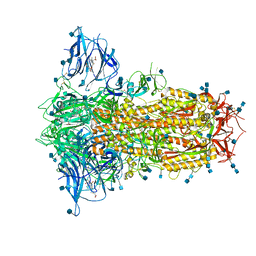 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Locked State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7A4N
 
 | | Cryo-EM structure of a prefusion stabilized SARS-CoV-2 Spike (D614N, R682S, R685G, A892P, A942P and V987P)(S-closed trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Rutten, L, Renault, L.L.R, Juraszek, J, Langedijk, J.P.M. | | Deposit date: | 2020-08-20 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Stabilizing the closed SARS-CoV-2 spike trimer.
Nat Commun, 12, 2021
|
|
