6UQK
 
 | | Cryo-EM structure of type 3 IP3 receptor revealing presence of a self-binding peptide | | Descriptor: | ZINC ION, inositol 1,4,5-triphosphate receptor, type 3 | | Authors: | Azumaya, C.M, Linton, E.A, Risener, C.J, Nakagawa, T, Karakas, E. | | Deposit date: | 2019-10-20 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Cryo-EM structure of human type-3 inositol triphosphate receptor reveals the presence of a self-binding peptide that acts as an antagonist.
J.Biol.Chem., 295, 2020
|
|
3TB4
 
 | | Crystal structure of the ISC domain of VibB | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, S, Zhang, C, Niu, B, Li, N, Liu, M, Wei, T, Zhu, D, Xu, S, Gu, L. | | Deposit date: | 2011-08-05 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insight into the ISC domain of VibB from Vibrio cholerae at atomic resolution: a snapshot just before the enzymatic reaction
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TG2
 
 | | Crystal structure of the ISC domain of VibB in complex with isochorismate | | Descriptor: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, TRIETHYLENE GLYCOL, Vibriobactin-specific isochorismatase | | Authors: | Liu, S, Zhang, C, Niu, B, Li, N, Liu, X, Liu, M, Wei, T, Zhu, D, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Structural insight into the ISC domain of VibB from Vibrio cholerae at atomic resolution: a snapshot just before the enzymatic reaction
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1A8M
 
 | | TUMOR NECROSIS FACTOR ALPHA, R31D MUTANT | | Descriptor: | TUMOR NECROSIS FACTOR ALPHA | | Authors: | Reed, C, Fu, Z.-Q, Wu, J, Xue, Y.-N, Harrison, R.W, Chen, M.-J, Weber, I.T. | | Deposit date: | 1998-03-27 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of TNF-alpha mutant R31D with greater affinity for receptor R1 compared with R2.
Protein Eng., 10, 1997
|
|
7NVR
 
 | | Human Mediator with RNA Polymerase II Pre-initiation complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Rengachari, S, Schilbach, S, Aibara, S, Cramer, P. | | Deposit date: | 2021-03-15 | | Release date: | 2021-05-05 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
1CPU
 
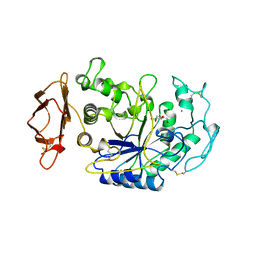 | | SUBSITE MAPPING OF THE ACTIVE SITE OF HUMAN PANCREATIC ALPHA-AMYLASE USING SUBSTRATES, THE PHARMACOLOGICAL INHIBITOR ACARBOSE, AND AN ACTIVE SITE VARIANT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, ... | | Authors: | Brayer, G.D, Sidhu, G, Maurus, R, Rydberg, E.H, Braun, C, Wang, Y, Nguyen, N.T, Overall, C.M, Withers, S.G. | | Deposit date: | 1999-06-07 | | Release date: | 1999-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subsite mapping of the human pancreatic alpha-amylase active site through structural, kinetic, and mutagenesis techniques.
Biochemistry, 39, 2000
|
|
4RF1
 
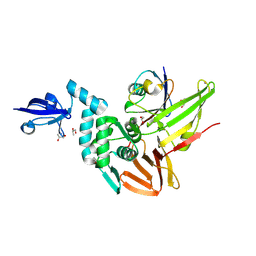 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease in complex with ubiquitin (space group P63) | | Descriptor: | 3-AMINOPROPANE, ORF1ab protein, S-1,2-PROPANEDIOL, ... | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
4REZ
 
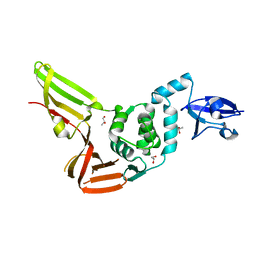 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease | | Descriptor: | ORF1ab protein, S-1,2-PROPANEDIOL, ZINC ION | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
4RF0
 
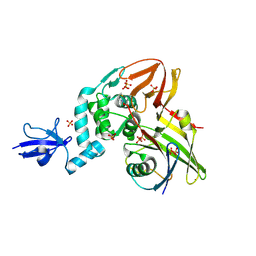 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease in complex with ubiquitin (space group P6522) | | Descriptor: | 3-AMINOPROPANE, ORF1ab protein, SULFATE ION, ... | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
1L6E
 
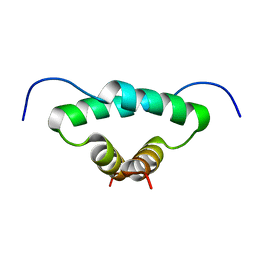 | | Solution structure of the docking and dimerization domain of protein kinase A II-alpha (RIIalpha D/D). Alternatively called the N-terminal dimerization domain of the regulatory subunit of protein kinase A. | | Descriptor: | cAMP-dependent protein kinase Type II-alpha regulatory chain | | Authors: | Morikis, D, Roy, M, Newlon, M.G, Scott, J.D, Jennings, P.A. | | Deposit date: | 2002-03-08 | | Release date: | 2002-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Electrostatic properties of the structure of the docking and dimerization domain of protein kinase A IIalpha
Eur.J.Biochem., 269, 2002
|
|
1ISO
 
 | |
2ECH
 
 | |
6QJI
 
 | |
6QJG
 
 | |
6QJK
 
 | |
6QJF
 
 | |
6QJL
 
 | |
6QJD
 
 | |
6QJN
 
 | |
3MKH
 
 | | Podospora anserina Nitroalkane Oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, NITROALKANE OXIDASE, ... | | Authors: | Tormos, J.R, Taylor, A.B, Daubner, S.C, Hart, P.J, Fitzpatrick, P.F. | | Deposit date: | 2010-04-14 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Identification of a hypothetical protein from Podospora anserina as a nitroalkane oxidase.
Biochemistry, 49, 2010
|
|
3CPU
 
 | | SUBSITE MAPPING OF THE ACTIVE SITE OF HUMAN PANCREATIC ALPHA-AMYLASE USING SUBSTRATES, THE PHARMACOLOGICAL INHIBITOR ACARBOSE, AND AN ACTIVE SITE VARIANT | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic alpha-amylase, ... | | Authors: | Brayer, G.D, Sidhu, G, Maurus, R, Rydberg, E.H, Braun, C, Wang, Y, Nguyen, N.T, Overall, C.M, Withers, S.G. | | Deposit date: | 1999-06-08 | | Release date: | 2001-06-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subsite mapping of the human pancreatic alpha-amylase active site through structural, kinetic, and mutagenesis techniques.
Biochemistry, 39, 2000
|
|
3MP2
 
 | |
6QJJ
 
 | |
1SPU
 
 | | STRUCTURE OF OXIDOREDUCTASE | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Wilmot, C.M, Phillips, S.E.V. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic mechanism of the quinoenzyme amine oxidase from Escherichia coli: exploring the reductive half-reaction.
Biochemistry, 36, 1997
|
|
5ZU2
 
 | | Effect of mutation (R554A) on FAD modification in Aspergillus oryzae RIB40formate oxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mikami, B, Uchida, H, Doubayashi, D. | | Deposit date: | 2018-05-06 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.441 Å) | | Cite: | The microenvironment surrounding FAD mediates its conversion to 8-formyl-FAD in Aspergillus oryzae RIB40 formate oxidase.
J.Biochem., 166, 2019
|
|
