7XVF
 
 | | Nav1.7 mutant class2 | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8CXJ
 
 | |
7XVE
 
 | | Human Nav1.7 mutant class-I | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8CWK
 
 | |
7XT6
 
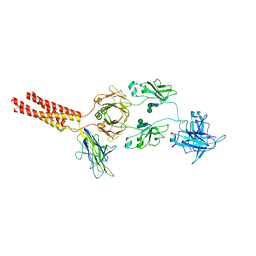 | | Structure of a membrane protein M3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain, ... | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cryo-EM structures of two human B cell receptor isotypes.
Science, 377, 2022
|
|
7XQ8
 
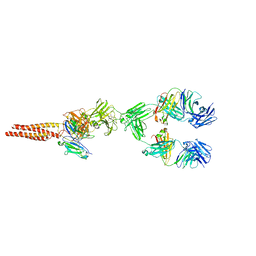 | | Structure of human B-cell antigen receptor of the IgM isotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, ... | | Authors: | Chen, M.Y, Su, Q, Shi, Y.G. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human IgM B cell receptor.
Science, 377, 2022
|
|
7UXO
 
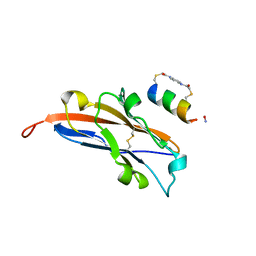 | | Structure of PDL1 in complex with FP30790, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, FP30790, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Travaline, T, Tattersfield, H, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXQ
 
 | | Structure of PDL1 in complex with FP28135, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, FP28135, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Travaline, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXP
 
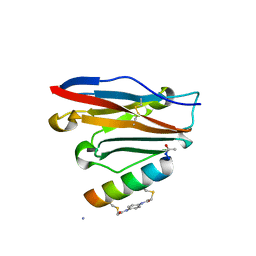 | | Structure of PDL1 in complex with FP28132, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, FP28132, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Travaline, T, Tattersfield, H, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UX5
 
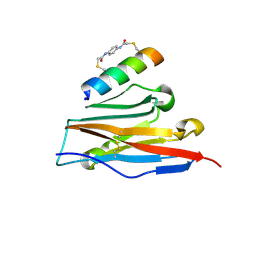 | | Structure of PDL1 in complex with FP28136, a Helicon Polypeptide | | Descriptor: | Helicon FP28136, N,N'-(1,4-phenylene)diacetamide, Programmed cell death 1 ligand 1 | | Authors: | Agarwal, S, Li, K, Tokareva, O, Thomson, T, Travaline, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UVF
 
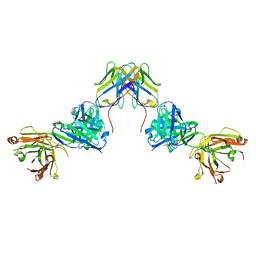 | | Crystal structure of ZED8 Fab complex with CD8 alpha | | Descriptor: | CHLORIDE ION, GLYCEROL, Immunoglobulin heavy chain, ... | | Authors: | Yu, C, Davies, C, Koerber, J.T, Williams, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Preclinical development of ZED8, an 89 Zr immuno-PET reagent for monitoring tumor CD8 status in patients undergoing cancer immunotherapy.
Eur J Nucl Med Mol Imaging, 50, 2023
|
|
7ZOZ
 
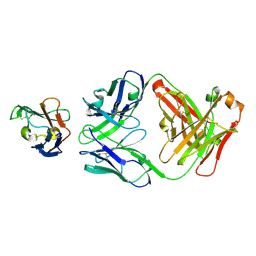 | | Crystal structure of Siglec-15 in complex with Fab | | Descriptor: | Anti-Siglec 15 Fab HC, Anti-Siglec 15 Fab LC, Sialic acid-binding Ig-like lectin 15 | | Authors: | Lenza, M.P, Oyenarte, I, Jimenez-Barbero, J, Ereno-Orbea, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural insights into Siglec-15 reveal glycosylation dependency for its interaction with T cells through integrin CD11b.
Nat Commun, 14, 2023
|
|
7XM9
 
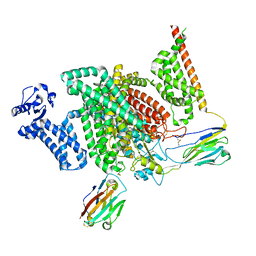 | | Cryo-EM structure of human NaV1.7/beta1/beta2-XEN907 | | Descriptor: | (7~{R})-1'-pentylspiro[6~{H}-furo[3,2-f][1,3]benzodioxole-7,3'-indole]-2'-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | zhang, J.T, Jiang, D.H. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XMF
 
 | | Cryo-EM structure of human NaV1.7/beta1/beta2-Nav1.7-IN2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[4-[3-(4-fluoranyl-2-methyl-phenoxy)azetidin-1-yl]pyrimidin-2-yl]amino]-~{N}-methyl-benzamide, ... | | Authors: | Zhang, J.T, Jiang, D.H. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XMG
 
 | | Cryo-EM structure of human NaV1.7/beta1/beta2-TCN-1752 | | Descriptor: | (1~{Z})-~{N}-[2-methyl-3-[(~{E})-[6-[4-[[4-(trifluoromethyloxy)phenyl]methoxy]piperidin-1-yl]-1~{H}-1,3,5-triazin-2-ylidene]amino]phenyl]ethanimidic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZLI
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with Dol25-P-Man and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLG
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with acceptor peptide and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Mao, R, Irobalieva, R, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLJ
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in ternary complex with Dol25-P-C-Man and acceptor peptide, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mao, R, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7UMG
 
 | | Crystal structure of human CD8aa-MR1-Ac-6-FP complex | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, Major histocompatibility complex class I-related gene protein, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2022-04-06 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD8 coreceptor engagement of MR1 enhances antigen responsiveness by human MAIT and other MR1-reactive T cells.
J.Exp.Med., 219, 2022
|
|
7XAE
 
 | | Crystal strucutre of PD-L1 and 3ONJA protein | | Descriptor: | 2IC6, Programmed cell death 1 ligand 1 | | Authors: | Liu, K.F, Xu, Z.P, Han, P, Gao, G.F, Chai, Y, Tan, S.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Crystal strucutre of PD-L1 and 2IC6 protein
To Be Published
|
|
7XAD
 
 | | Crystal strucutre of PD-L1 and DBL2_02 designed protein binder | | Descriptor: | DBL2_02 binder, Programmed cell death 1 ligand 1 | | Authors: | Liu, K.F, Xu, Z.P, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7TY0
 
 | |
7WVM
 
 | | The complex structure of PD-1 and cemiplimab | | Descriptor: | Heavy Chain of Cemiplimab, Light Chain of Cemiplimab, Programmed cell death protein 1 | | Authors: | Lu, D, Xu, Z.P, Liu, K.F, Tan, S.G, Gao, G.F, Chai, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | PD-1 N58-Glycosylation-Dependent Binding of Monoclonal Antibody Cemiplimab for Immune Checkpoint Therapy.
Front Immunol, 13, 2022
|
|
7WSL
 
 | | PD-1 in complex with Dostarlimab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2022-01-30 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Molecular basis of PD-1 blockade by dostarlimab, the FDA-approved antibody for cancer immunotherapy.
Biochem.Biophys.Res.Commun., 599, 2022
|
|
7WSO
 
 | | Structure of a membrane protein G | | Descriptor: | B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, Immunoglobulin heavy constant gamma 1 | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-01-30 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM structures of two human B cell receptor isotypes.
Science, 377, 2022
|
|
