7F8N
 
 | | Human pannexin-1 showing a conformational change in the N-terminal domain and blocked pore | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
8Q0J
 
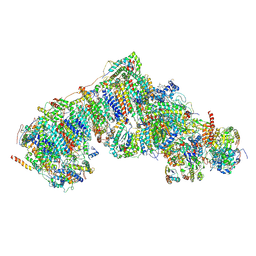 | | Inward-facing, slack proteoliposome complex I at 3.8 A. Initially purified in DDM. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
8Q48
 
 | | Outward-facing, closed proteoliposome complex I at 2.5 A. Initially purified in LMNG. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-08-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
7FJ2
 
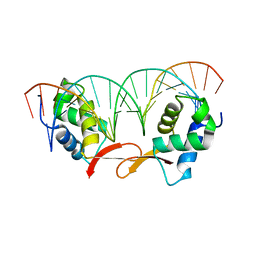 | | Structure of FOXM1 homodimer bound to a palindromic DNA site | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*AP*AP*AP*CP*AP*TP*GP*TP*TP*TP*AP*CP*GP*GP*T)-3'), Forkhead box protein M1 | | Authors: | Dai, S.Y, Li, J, Zhang, H.J. | | Deposit date: | 2021-08-02 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Mechanistic Insights into the Preference for Tandem Binding Sites in DNA Recognition by FOXM1.
J.Mol.Biol., 434, 2021
|
|
7Q6O
 
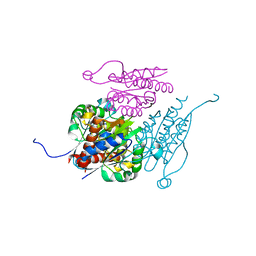 | | Structure of WrbA from Yersinia pseudotuberculosis in C2221 | | Descriptor: | CHLORIDE ION, NAD(P)H dehydrogenase (quinone) | | Authors: | Gabrielsen, M, Beckham, K.S.H, Roe, A.J. | | Deposit date: | 2021-11-08 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of WrbA, a spurious target of the salicylidene acylhydrazide inhibitors of type III secretion in Gram-negative pathogens, and verification of improved specificity of next-generation compounds.
Microbiology (Reading, Engl.), 168, 2022
|
|
8Q0M
 
 | | Outward-facing, closed proteoliposome complex I at 3.1 A. Initially purified in DDM. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
8Q1Y
 
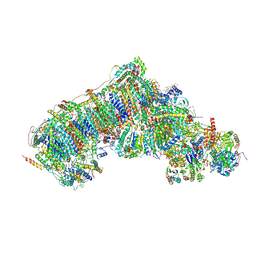 | | Outward-facing, open2 proteoliposome complex I at 2.6 A, after deactivation treatment. Initially purified in LMNG. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
8Q1P
 
 | | Inward-facing, open2 proteoliposome complex I at 2.9 A, after deactivation treatment. Initially purified in LMNG. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
7QBA
 
 | | CryoEM structure of the ABC transporter NosDFY complexed with nitrous oxide reductase NosZ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zipfel, S, Mueller, C, Topitsch, A, Lutz, M, Zhang, L, Einsle, O. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
8Q1U
 
 | | Inward-facing, open1 proteoliposome complex I at 3.3 A, after deactivation treatment. Initially purified in LMNG. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
4FQX
 
 | | Crystal structure of HLA-DM bound to HLA-DR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DM alpha chain, ... | | Authors: | Sethi, D.K, Pos, W, Wucherpfennig, K.W. | | Deposit date: | 2012-06-25 | | Release date: | 2013-01-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Crystal Structure of the HLA-DM-HLA-DR1 Complex Defines Mechanisms for Rapid Peptide Selection.
Cell(Cambridge,Mass.), 151, 2012
|
|
7Q6N
 
 | | Structure of WrbA from Salmonella Typhimurium bound to ME0052 | | Descriptor: | 2-azanyl-4,6-bis(bromanyl)phenol, FLAVIN MONONUCLEOTIDE, NAD(P)H dehydrogenase (quinone), ... | | Authors: | Gabrielsen, M, Beckham, K.S.H, Roe, A.J. | | Deposit date: | 2021-11-08 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of WrbA, a spurious target of the salicylidene acylhydrazide inhibitors of type III secretion in Gram-negative pathogens, and verification of improved specificity of next-generation compounds.
Microbiology (Reading, Engl.), 168, 2022
|
|
8Q25
 
 | | Outward-facing, open1 proteoliposome complex I at 2.8 A, after deactivation treatment. Initially purified in LMNG. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I.
Science, 384, 2024
|
|
7BXU
 
 | | CLC-7/Ostm1 membrane protein complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H(+)/Cl(-) exchange transporter 7, Osteopetrosis-associated transmembrane protein 1 | | Authors: | Zhang, S.S, Yang, M.J. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular insights into the human CLC-7/Ostm1 transporter.
Sci Adv, 6, 2020
|
|
7Q6M
 
 | | Structure of WrbA from Yersinia pseudotuberculosis in P1 | | Descriptor: | CHLORIDE ION, NAD(P)H dehydrogenase (quinone) | | Authors: | Gabrielsen, M, Beckham, K.S.H, Roe, A.J. | | Deposit date: | 2021-11-08 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structures of WrbA, a spurious target of the salicylidene acylhydrazide inhibitors of type III secretion in Gram-negative pathogens, and verification of improved specificity of next-generation compounds.
Microbiology (Reading, Engl.), 168, 2022
|
|
7QVH
 
 | |
7BYX
 
 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A E208A with beta-1,3-galactotriose | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
8Q74
 
 | |
8PJE
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting influenza A virus haemagglutinin (HA)306-318 PKYVKQNTLKLAT | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | MacLachlan, B.J, Wall, A, Greenshields-Watson, A.L, Hesketh, S.J, Cole, D.K, Rizkallah, P.J, Godkin, A.J. | | Deposit date: | 2023-06-23 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A targeted single mutation in influenza A virus universal epitope transforms immunogenicity and protective immunity via CD4 + T cell activation.
Cell Rep, 43, 2024
|
|
7F6V
 
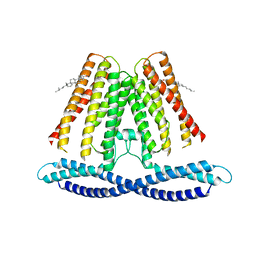 | | Cryo-EM structure of the human TACAN channel in a closed state | | Descriptor: | CHOLESTEROL, Ion channel TACAN | | Authors: | Chen, X.Z, Wang, Y.J, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2021-06-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structure of the human TACAN in a closed state.
Cell Rep, 38, 2022
|
|
4FDC
 
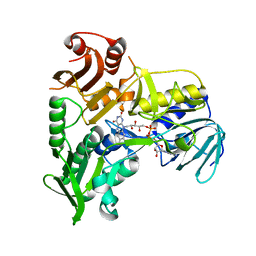 | |
8PEM
 
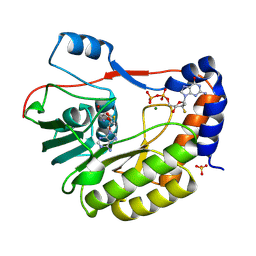 | |
8PJF
 
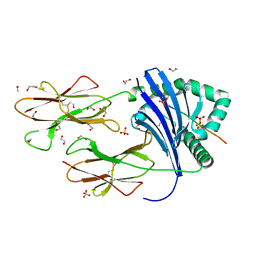 | | Human Leukocyte Antigen class II allotype DR1 presenting P11T->R modified influenza A virus haemagglutinin (HA)306-318 PKYVKQNTLKLAR | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | MacLachlan, B.J, Wall, A, Greenshields-Watson, A.L, Cole, D.K, Rizkallah, P.J, Godkin, A.J. | | Deposit date: | 2023-06-23 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A targeted single mutation in influenza A virus universal epitope transforms immunogenicity and protective immunity via CD4 + T cell activation.
Cell Rep, 43, 2024
|
|
7F0M
 
 | | Crystal Structure of human Pin1 complexed with a potent covalent inhibitor | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 8-(2-chloranylethanoyl)-4-[(5-naphthalen-1-ylfuran-2-yl)methyl]-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7Q7F
 
 | | Room temperature structure of the Rhodobacter Sphaeroides Photosynthetic Reaction Center F(M197)H mutant at atmospheric pressure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Selikhanov, G.K, Gabdulkhakov, A.G, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
