8Q3C
 
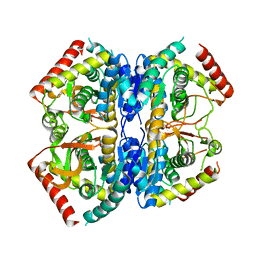 | | Structure of Selenomonas ruminantium lactate dehydrogenase I85R mutant | | Descriptor: | CHLORIDE ION, L-lactate dehydrogenase, NITRATE ION, ... | | Authors: | Bertrand, Q, Coquille, S, Iorio, A, Sterpone, F, Madern, D. | | Deposit date: | 2023-08-03 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete allosteric regulation acquisition in the super family of lactate and malate dehydrogenases.
J.Struct.Biol., 215, 2023
|
|
8TO0
 
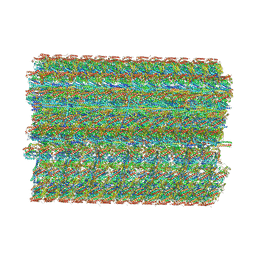 | | 48-nm repeating structure of doublets from mouse sperm flagella | | Descriptor: | Cilia- and flagella- associated protein 210, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Chen, Z, Shiozak, M, Hass, K.M, Skinner, W, Zhao, S, Guo, C, Polacco, B.J, Yu, Z, Krogan, N.J, Kaake, R.M, Vale, R.D, Agard, D.A. | | Deposit date: | 2023-08-02 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | De novo protein identification in mammalian sperm using in situ cryoelectron tomography and AlphaFold2 docking.
Cell, 186, 2023
|
|
8TNK
 
 | |
8TNR
 
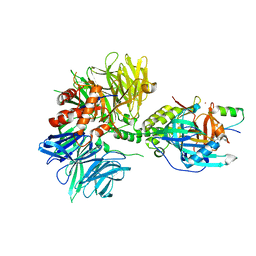 | | Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 2 | | Descriptor: | 2-[(3S)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1H-isoindole-1,3(2H)-dione, DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
8TNP
 
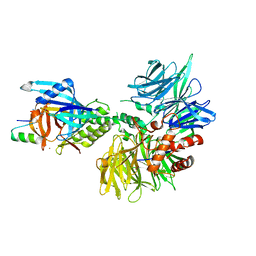 | | Cryo-EM structure of DDB1dB:CRBN:Pomalidomide:SD40 | | Descriptor: | DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, Protein cereblon, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
8TNQ
 
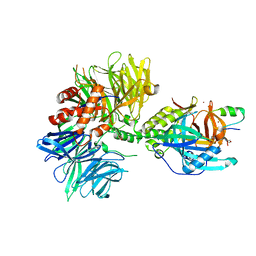 | | Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 1 | | Descriptor: | 2-[(3S)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1H-isoindole-1,3(2H)-dione, DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
8Q1Z
 
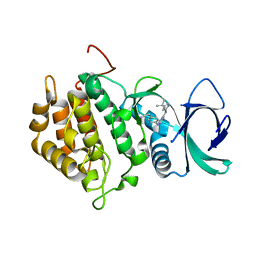 | |
8K9K
 
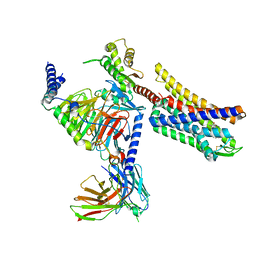 | | Full agonist-bound mu-type opioid receptor-G protein complex | | Descriptor: | DAMGO, G protein subunit alpha i3, Guanine nucleotide binding protein, ... | | Authors: | Hisano, T, Uchikubo-Kamo, T, Shirouzu, M, Imai, S, Kaneko, S, Shimada, I. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural and dynamic insights into the activation of the mu-opioid receptor by an allosteric modulator.
Nat Commun, 15, 2024
|
|
8K9L
 
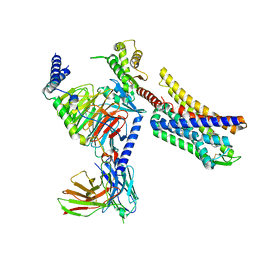 | | Full agonist- and positive allosteric modulator-bound mu-type opioid receptor-G protein complex | | Descriptor: | (2~{S})-2-(3-bromanyl-4-methoxy-phenyl)-3-(4-chlorophenyl)sulfonyl-1,3-thiazolidine, DAMGO, G protein subunit alpha i3, ... | | Authors: | Hisano, T, Uchikubo-Kamo, T, Shirouzu, M, Imai, S, Kaneko, S, Shimada, I. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural and dynamic insights into the activation of the mu-opioid receptor by an allosteric modulator.
Nat Commun, 15, 2024
|
|
8Q1B
 
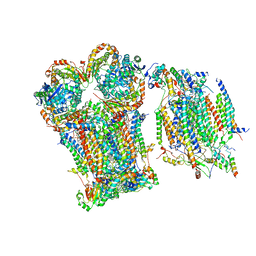 | | III2-IV1 respiratory supercomplex from S. pombe | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Moe, A, Brzezinski, P. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of the S. pombe III-IV-cyt c supercomplex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8K8M
 
 | |
8K8J
 
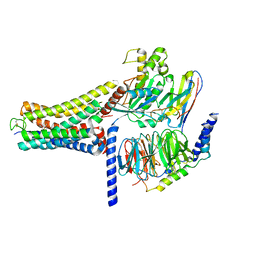 | | Cannabinoid Receptor 1 bound to Fenofibrate coupling MiniGsq and Nb35 Complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Tang, W.Q, Wang, T.X, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-07-30 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Fenofibrate Recognition and G q Protein Coupling Mechanisms of the Human Cannabinoid Receptor CB1.
Adv Sci, 11, 2024
|
|
8Q0V
 
 | | Endothiapepsin in complex with ligand (3R,5R)-3-(2-((methyl(prop-2-yn-1-yl)amino)methyl)thiazol-4-yl)-5-(3-(4-(trifluoromethyl)phenyl)-1,2,4-oxadiazol-5-yl)pyrrolidin-3-ol (CBWS-SE-073) | | Descriptor: | (3~{R},5~{R})-3-[2-[[methyl(prop-2-ynyl)amino]methyl]-1,3-thiazol-4-yl]-5-[3-[4-(trifluoromethyl)phenyl]-1,2,4-oxadiazol-5-yl]pyrrolidin-3-ol, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
8Q0W
 
 | | Endothiapepsin in complex with ligand (3R,5R)-3-(2-((allyl(methyl)amino)methyl)thiazol-4-yl)-5-(3-(4-(trifluoromethyl)phenyl)-1,2,4-oxadiazol-5-yl)pyrrolidin-3-ol (CBWS-SE-086) | | Descriptor: | (3~{R},5~{R})-3-[2-[[methyl(prop-2-enyl)amino]methyl]-1,3-thiazol-4-yl]-5-[3-[4-(trifluoromethyl)phenyl]-1,2,4-oxadiazol-5-yl]pyrrolidin-3-ol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
8Q10
 
 | | Endothiapepsin in complex with ligand (3R,5R)-3-(2-(hydroxymethyl)thiazol-4-yl)-5-(3-(4-(trifluoromethyl)phenyl)-1,2,4-oxadiazol-5-yl)pyrrolidin-3-ol (CBWS-SE-125) | | Descriptor: | (3~{R},5~{R})-3-[2-(hydroxymethyl)-1,3-thiazol-4-yl]-5-[3-[4-(trifluoromethyl)phenyl]-1,2,4-oxadiazol-5-yl]pyrrolidin-3-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
8Q12
 
 | | Endothiapepsin in complex with ligand (3R,5R)-5-(3-(2-aminophenyl)-1,2,4-oxadiazol-5-yl)-3-(2-((methyl(prop-2-yn-1-yl)amino)methyl)thiazol-4-yl)pyrrolidin-3-ol (CBWS-SE-146.2) | | Descriptor: | (3~{R},5~{R})-5-[3-(2-aminophenyl)-1,2,4-oxadiazol-5-yl]-3-[2-[[methyl(prop-2-ynyl)amino]methyl]-1,3-thiazol-4-yl]pyrrolidin-3-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
8Q14
 
 | | Endothiapepsin in complex with ligand (3R,5R)-3-(2-((methyl(prop-2-yn-1-yl)amino)methyl)thiazol-4-yl)-5-(3-(2-nitro-4-(trifluoromethyl)phenyl)-1,2,4-oxadiazol-5-yl)pyrrolidin-3-ol (CBWS-SE-171) | | Descriptor: | (3~{R},5~{R})-3-[2-[[methyl(prop-2-ynyl)amino]methyl]-1,3-thiazol-4-yl]-5-[3-[2-nitro-4-(trifluoromethyl)phenyl]-1,2,4-oxadiazol-5-yl]pyrrolidin-3-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
8Q13
 
 | | Endothiapepsin in complex with ligand (3R,5R)-5-(3-(2-amino-4-(trifluoromethyl)phenyl)-1,2,4-oxadiazol-5-yl)-3-(2-((methyl(prop-2-yn-1-yl)amino)methyl)thiazol-4-yl)pyrrolidin-3-ol (CBWS-SE-168) | | Descriptor: | (3~{R},5~{R})-5-[3-[2-azanyl-4-(trifluoromethyl)phenyl]-1,2,4-oxadiazol-5-yl]-3-[2-[[methyl(prop-2-ynyl)amino]methyl]-1,3-thiazol-4-yl]pyrrolidin-3-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
8Q11
 
 | | Endothiapepsin in complex with ligand (3R,5R)-5-(3-(4-fluorophenyl)-1,2,4-oxadiazol-5-yl)-3-(2-((methyl(prop-2-yn-1-yl)amino)methyl)thiazol-4-yl)pyrrolidin-3-ol (CBWS-SE-126) | | Descriptor: | (3~{R},5~{R})-5-[3-(4-fluorophenyl)-1,2,4-oxadiazol-5-yl]-3-[2-[[methyl(prop-2-ynyl)amino]methyl]-1,3-thiazol-4-yl]pyrrolidin-3-ol, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
8Q0X
 
 | | Endothiapepsin in complex with ligand (3R,5R)-3-(2-((methyl(propyl)amino)methyl)thiazol-4-yl)-5-(3-(4-(trifluoromethyl)phenyl)-1,2,4-oxadiazol-5-yl)pyrrolidin-3-ol (CBWS-SE-087) | | Descriptor: | (3~{R},5~{R})-3-[2-[[methyl(propyl)amino]methyl]-1,3-thiazol-4-yl]-5-[3-[4-(trifluoromethyl)phenyl]-1,2,4-oxadiazol-5-yl]pyrrolidin-3-ol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
8Q0Y
 
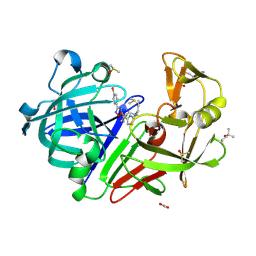 | | Endothiapepsin in complex with ligand (3R,5R)-5-(3-(4-bromophenyl)-1,2,4-oxadiazol-5-yl)-3-(2-((methyl(prop-2-yn-1-yl)amino)methyl)thiazol-4-yl)pyrrolidin-3-ol (CBWS-SE-085.1) | | Descriptor: | (3~{R},5~{R})-5-[3-(4-bromophenyl)-1,2,4-oxadiazol-5-yl]-3-[2-[[methyl(prop-2-ynyl)amino]methyl]-1,3-thiazol-4-yl]pyrrolidin-3-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
8Q0Z
 
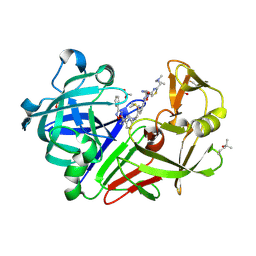 | | Endothiapepsin in complex with ligand (3R,5R)-3-(2-((methyl(prop-2-yn-1-yl)amino)methyl)thiazol-4-yl)-5-(3-phenyl-1,2,4-oxadiazol-5-yl)pyrrolidin-3-ol (CBWS-SE-089) | | Descriptor: | (3~{R},5~{R})-3-[2-[[methyl(prop-2-ynyl)amino]methyl]-1,3-thiazol-4-yl]-5-(3-phenyl-1,2,4-oxadiazol-5-yl)pyrrolidin-3-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
8K8H
 
 | |
8Q0N
 
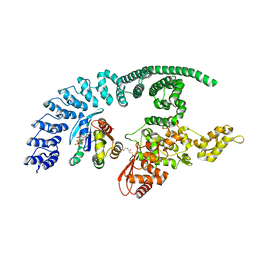 | | HACE1 in complex with RAC1 Q61L | | Descriptor: | E3 ubiquitin-protein ligase HACE1, GUANOSINE-5'-TRIPHOSPHATE, Ras-related C3 botulinum toxin substrate 1, ... | | Authors: | Wolter, M, Duering, J, Dienemann, C, Lorenz, S. | | Deposit date: | 2023-07-28 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8TLM
 
 | | Structure of a class A GPCR/Fab complex | | Descriptor: | C-C chemokine receptor type 8, Green fluorescent protein fusion, Fab heavy chain, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
