5BS8
 
 | | Crystal structure of a topoisomerase II complex | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | Deposit date: | 2015-06-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6MBQ
 
 | | Crystal structure of Mg-free wild-type KRAS (2-166) bound to GMPPNP in the state 1 conformation | | Descriptor: | 1,2-ETHANEDIOL, GTPase KRas, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Dharmaiah, S, Davies, D.R, Abendroth, J, Gillette, W.G, Stephen, A.G, Simanshu, D.K. | | Deposit date: | 2018-08-30 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of N-terminally processed KRAS provide insight into the role of N-acetylation.
Sci Rep, 9, 2019
|
|
6N0Q
 
 | | BRAF in complex with N-(4-methyl-3-(1-methyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)phenyl)-3-(trifluoromethyl)benzamide. | | Descriptor: | N-[4-methyl-3-(1-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)phenyl]-3-(trifluoromethyl)benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-11-07 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Design and Discovery ofN-(3-(2-(2-Hydroxyethoxy)-6-morpholinopyridin-4-yl)-4-methylphenyl)-2-(trifluoromethyl)isonicotinamide, a Selective, Efficacious, and Well-Tolerated RAF Inhibitor Targeting RAS Mutant Cancers: The Path to the Clinic.
J.Med.Chem., 63, 2020
|
|
1BVJ
 
 | | HIV-1 RNA A-RICH HAIRPIN LOOP | | Descriptor: | RNA (5'-R(P*GP*GP*CP*GP*AP*CP*GP*GP*UP*GP*UP*AP*AP*AP*AP*AP*UP*CP*UP*CP*GP*CP* C)-3') | | Authors: | Puglisi, E.V, Puglisi, J.D. | | Deposit date: | 1998-08-31 | | Release date: | 1998-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HIV-1 A-rich RNA loop mimics the tRNA anticodon structure.
Nat.Struct.Biol., 5, 1998
|
|
6FOL
 
 | | Domain II of the human copper chaperone in complex with human Cu,Zn superoxide dismutase | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Copper chaperone for superoxide dismutase, GLYCEROL, ... | | Authors: | Sala, F.A, Wright, G.S.A, Antonyuk, S.V, Garratt, R.C, Hasnain, S.S. | | Deposit date: | 2018-02-07 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular recognition and maturation of SOD1 by its evolutionarily destabilised cognate chaperone hCCS.
Plos Biol., 17, 2019
|
|
6S57
 
 | |
3K1U
 
 | | Beta-xylosidase, family 43 glycosyl hydrolase from Clostridium acetobutylicum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-xylosidase, ... | | Authors: | Osipiuk, J, Wu, R, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystal structure of
beta-xylosidase, family 43 glycosyl hydrolase from Clostridium acetobutylicum at 1.55 A resolution
To be Published
|
|
4YVF
 
 | | Structure of S-adenosyl-L-homocysteine hydrolase | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{[5-chloro-2-(4-chlorophenoxy)phenyl](2-{[2-(methylamino)ethyl]amino}-2-oxoethyl)amino}-N-(1,3-dihydro-2H-isoindol-2-yl)-N-methylacetamide, Adenosylhomocysteinase | | Authors: | Akiko, K. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and structural analyses of S-adenosyl-L-homocysteine hydrolase inhibitors based on non-adenosine analogs.
Bioorg.Med.Chem., 23, 2015
|
|
2QA7
 
 | |
6D3X
 
 | |
5FRM
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ384 (compound 4a) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-azanylidene-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-1,8-naphthyridine-3-carboxamide, 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP *TP*CP*GP*CP*A)-3', ... | | Authors: | Maskell, D.P, Pye, V.E, Cherepanov, P. | | Deposit date: | 2015-12-18 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | HIV-1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases.
Acs Chem.Biol., 11, 2016
|
|
3WTM
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid | | Descriptor: | 2-chloro-5-{[(2E)-2-(nitromethylidene)imidazolidin-1-yl]methyl}pyridine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3K6T
 
 | | Crystal structure of the GLD-1 homodimerization domain from Caenorhabditis elegans at 2.04 A resolution | | Descriptor: | Female germline-specific tumor suppressor gld-1 | | Authors: | Beuck, C, Szymczyna, B.R, Kerkow, D.E, Carmel, A.B, Columbus, L, Stanfield, R.L, Williamson, J.R. | | Deposit date: | 2009-10-09 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of the GLD-1 homodimerization domain: insights into STAR protein-mediated translational regulation.
Structure, 18, 2010
|
|
6GAK
 
 | |
3KBL
 
 | | Crystal structure of the GLD-1 homodimerization domain from Caenorhabditis elegans N169A mutant at 2.28 A resolution | | Descriptor: | Female germline-specific tumor suppressor gld-1 | | Authors: | Beuck, C, Szymczyna, B.R, Kerkow, D.E, Carmel, A.B, Columbus, L, Stanfield, R.L, Williamson, J.R. | | Deposit date: | 2009-10-20 | | Release date: | 2010-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of the GLD-1 homodimerization domain: insights into STAR protein-mediated translational regulation.
Structure, 18, 2010
|
|
3KE2
 
 | |
5MMD
 
 | | TMB-1. Structural insights into TMB-1 and the role of residue 119 and 228 in substrate and inhibitor binding | | Descriptor: | CHLORIDE ION, Metallo-beta-lactamase 1, ZINC ION | | Authors: | Skagseth, S, Christopeit, T, Akhter, S, Bayer, A, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2016-12-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights into TMB-1 and the Role of Residues 119 and 228 in Substrate and Inhibitor Binding.
Antimicrob. Agents Chemother., 61, 2017
|
|
5FUM
 
 | | Mus musculus acetylcholinesterase in complex with AL200 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-(biphenyl-4-yloxy)-1-[4-(4-ethylpiperazin-1-yl)piperidin-1-yl]ethanone, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Knutsson, S, Engdahl, C, Ekstrom, F, Linusson, A. | | Deposit date: | 2016-01-28 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Selective Inhibitors Targeting Acetylcholinesterase 1 from Disease-Transmitting Mosquitoes.
J.Med.Chem., 17, 2016
|
|
3WTL
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein Complexed with Nitromethylene Analogue of Imidacloprid | | Descriptor: | 2-chloro-5-{[(2E)-2-(nitromethylidene)imidazolidin-1-yl]methyl}pyridine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
5G2T
 
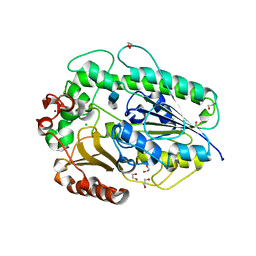 | | BT1596 in complex with its substrate 4,5 unsaturated uronic acid alpha 1,4 D-Glucosamine-2-N, 6-O-disulfate | | Descriptor: | 1,2-ETHANEDIOL, 2-O GLYCOSAMINOGLYCAN SULFATASE, 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid, ... | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3KHI
 
 | |
6S56
 
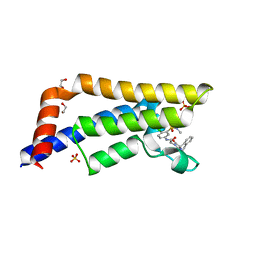 | | Crystal structure of human ATAD2 bromodomain in complex with N-(4-chloro-3-(N,N-dimethylsulfamoyl)phenyl)-2-(2,5-dioxo-3',4'-dihydro-2'H-spiro[imidazolidine-4,1'-naphthalen]-1-yl)acetamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4~{R})-2',5'-bis(oxidanylidene)spiro[2,3-dihydro-1~{H}-naphthalene-4,4'-imidazolidine]-1'-yl]-~{N}-[4-chloranyl-3-(dimethylsulfamoyl)phenyl]ethanamide, ATPase family AAA domain-containing protein 2, ... | | Authors: | Chung, C. | | Deposit date: | 2019-06-30 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A Qualified Success: Discovery of a New Series of ATAD2 Bromodomain Inhibitors with a Novel Binding Mode Using High-Throughput Screening and Hit Qualification.
J.Med.Chem., 62, 2019
|
|
7WLG
 
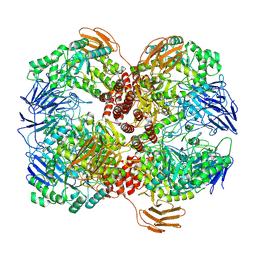 | | Cryo-EM structure of GH31 alpha-1,3-glucosidase from Lactococcus lactis subsp. cremoris | | Descriptor: | Alpha-xylosidase | | Authors: | Ikegaya, M, Moriya, T, Adachi, N, Kawasaki, M, Park, E.Y, Miyazaki, T. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
8GX3
 
 | | The crystal structure of human Calpain-1 protease core in complex with 14c | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, N-[(2S)-3-cyclohexyl-1-[[(2S,3S)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of human Calpain-1 protease core in complex with 14c
To Be Published
|
|
5BX1
 
 | | Crystal Structure of PRL-1 complex with compound analogy 3 | | Descriptor: | 3-(5,6-dimethyl-2H-isoindol-2-yl)-N'-[(E)-furan-2-ylmethylidene]benzohydrazide, Protein tyrosine phosphatase type IVA 1, SULFATE ION | | Authors: | Liu, S, Bai, Y, Zhang, Z. | | Deposit date: | 2015-06-08 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of PRL-1 complex with compound analogy 3
To Be Published
|
|
