6R55
 
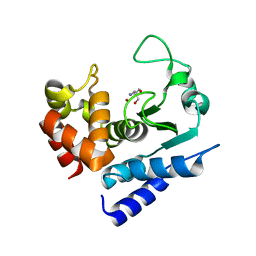 | | Crystal structure of PPEP-1(E184K) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ZINC ION | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2019-03-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
4UNV
 
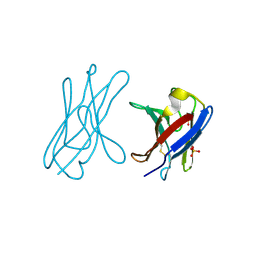 | | Covalent dimer of lambda variable domains | | Descriptor: | IG LAMBDA CHAIN V-II REGION MGC, SULFATE ION | | Authors: | Brumshtein, B, Esswein, S, Landau, M, Ryan, C, Whitelegge, J, Sawaya, M, Eisenberg, D.S. | | Deposit date: | 2014-05-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Formation of Amyloid Fibers by Monomeric Light-Chain Variable Domains.
J.Biol.Chem., 289, 2014
|
|
6RBE
 
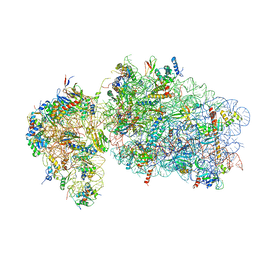 | | State 2 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
4P3G
 
 | |
6R5I
 
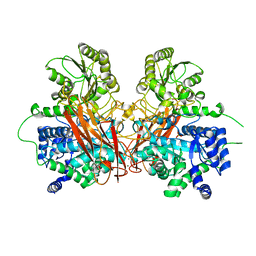 | |
4UJ8
 
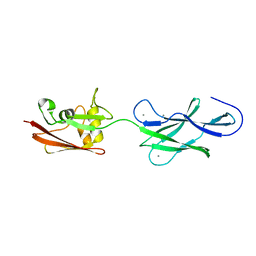 | | Structure of surface layer protein SbsC, domains 6-7 | | Descriptor: | CALCIUM ION, SURFACE LAYER PROTEIN | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Surface Layer Protein Sbsc
To be Published
|
|
6R5Y
 
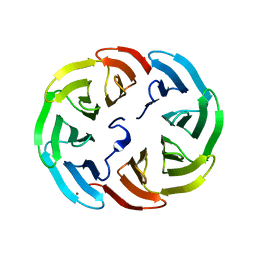 | |
4UOO
 
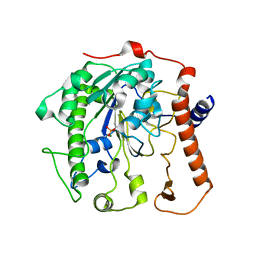 | |
4UGS
 
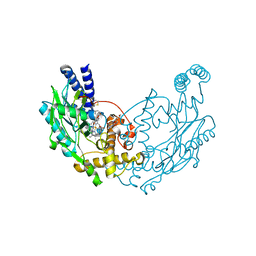 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with N,N'-(ethane-1,2-diylbis(oxybenzene-3,1-diyl))dithiophene-2- carboximidamide | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORIDE ION, N,N'-[ethane-1,2-diylbis(oxybenzene-3,1-diyl)]dithiophene-2-carboximidamide, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
6R7M
 
 | | Tobacco Mosaic Virus (TMV) | | Descriptor: | Capsid protein | | Authors: | Schmidli, C, Albiez, S, Rima, L, Righetto, R, Mohammed, I, Oliva, P, Kovacik, L, Stahlberg, H, Braun, T. | | Deposit date: | 2019-03-29 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (1.92 Å) | | Cite: | Microfluidic protein isolation and sample preparation for high-resolution cryo-EM.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4P4C
 
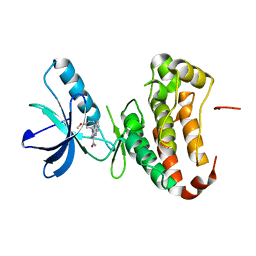 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-(3-methoxyphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
4UHL
 
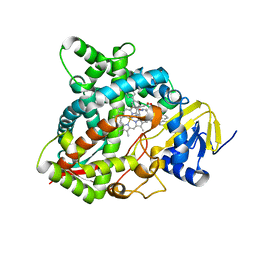 | | HUMAN STEROL 14-ALPHA DEMETHYLASE (CYP51) IN COMPLEX WITH VFV IN P1 SPACE GROUP | | Descriptor: | N-[(1R)-1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, I Lepesheva, G. | | Deposit date: | 2015-03-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human Sterol 14Alpha-Demethylase (Cyp51) as a Target for Anticancer Chemotherapy: Towards Structure-Aided Drug Design.
J.Lipid Res., 57, 2016
|
|
4UIG
 
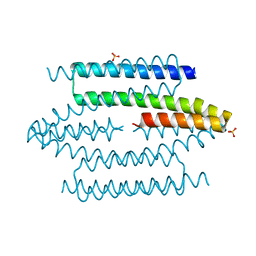 | |
6R8N
 
 | | STRUCTURE DETERMINATION OF THE TETRAHEDRAL AMINOPEPTIDASE TET2 FROM P. HORIKOSHII BY USE OF COMBINED SOLID-STATE NMR, SOLUTION-STATE NMR AND EM DATA 4.1 A, FOLLOWED BY REAL_SPACE_REFINEMENT AT 4.1 A | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Colletier, J.-P, Gauto, D, Estrozi, L, Favier, A, Effantin, G, Schoehn, G, Boisbouvier, J, Schanda, P. | | Deposit date: | 2019-04-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
4UP0
 
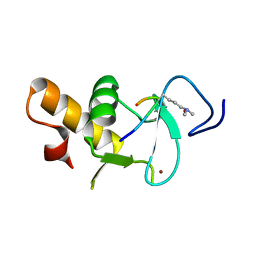 | | Ternary crystal structure of the Pygo2 PHD finger in complex with the B9L HD1 domain and a H3K4me2 peptide | | Descriptor: | HISTONE H3.1, PYGOPUS HOMOLOG 2, B-CELL CLL/LYMPHOMA 9-LIKE PROTEIN, ... | | Authors: | Miller, T.C.R, Fiedler, M, Rutherford, T.J, Birchall, K, Chugh, J, Bienz, M. | | Deposit date: | 2014-06-11 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Competitive Binding of a Benzimidazole to the Histone-Binding Pocket of the Pygo Phd Finger.
Acs Chem.Biol., 9, 2014
|
|
4UPI
 
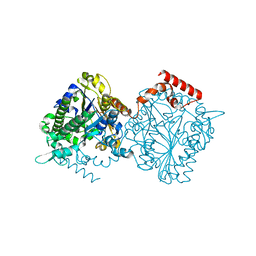 | | Dimeric sulfatase SpAS1 from Silicibacter pomeroyi | | Descriptor: | SULFATASE FAMILY PROTEIN, ZINC ION | | Authors: | Jonas, S, van Loo, B, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2014-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Balancing Specificity and Promiscuity in Enzyme Evolution: Multidimensional Activity Transitions in the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 141, 2019
|
|
4UOG
 
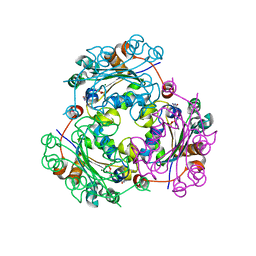 | | Crystallographic structure of nucleoside diphosphate kinase from Litopenaeus vannamei complexed with dCDP | | Descriptor: | DEOXYCYTIDINE DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Lopez-Zavala, A.A, Stojanoff, V, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2014-06-03 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Nucleoside Diphosphate Kinase from Pacific Shrimp (Litopenaeus Vannamei) in Binary Complexes with Purine and Pyrimidine Nucleoside Diphosphates
Acta Crystallogr.,Sect.F, 79, 2014
|
|
4PEQ
 
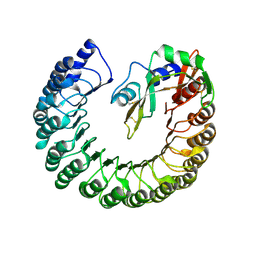 | | Structure of bovine ribonuclease inhibitor complexed with bovine ribonuclease I | | Descriptor: | Ribonuclease pancreatic, Ribonuclease/angiogenin inhibitor 1 | | Authors: | Bianchetti, C.M, Lomax, J.E, Raines, R.T, Fox, B.G. | | Deposit date: | 2014-04-24 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Functional evolution of ribonuclease inhibitor: insights from birds and reptiles.
J.Mol.Biol., 426, 2014
|
|
6R3Q
 
 | |
4UJ9
 
 | | Protein Kinase A in complex with an Inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[(3S,4R)-4-[4-(trifluoromethyl)phenyl]carbonylpyrrolidin-3-yl]-3H-quinazolin-4-one, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Alam, K.A, Engh, R.A. | | Deposit date: | 2015-04-09 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Addressing the Glycine-Rich Loop of Protein Kinases by a Multi-Facetted Interaction Network: Inhibition of Pka and a Pkb Mimic.
Chemistry, 22, 2016
|
|
4P6Z
 
 | |
4UPV
 
 | | Low X-ray dose structure of a Ni-A Ni-Sox mixture of the D. fructosovorans NiFe-hydrogenase L122A mutant | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-18 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
4P7E
 
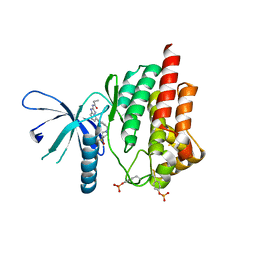 | | Triazolopyridine compounds as selective JAK1 inhibitors: from hit identification to GLPG0634 | | Descriptor: | N-(5-{4-[(1,1-dioxidothiomorpholin-4-yl)methyl]phenyl}[1,2,4]triazolo[1,5-a]pyridin-2-yl)cyclopropanecarboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Menet, C.C.J, Fletcher, S, Van Lommen, G, Geney, R, Blanc, J, Smits, K, Jouannigot, N, van der Aar, E.M, Clement-Lacroix, P, Lepescheux, L, Galien, R, Vayssiere, B, Nelles, L, Christophe, T, Brys, R, Uhring, M, Ciesielski, F, Van Rompaey, L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Triazolopyridines as Selective JAK1 Inhibitors: From Hit Identification to GLPG0634.
J.Med.Chem., 57, 2014
|
|
6RA1
 
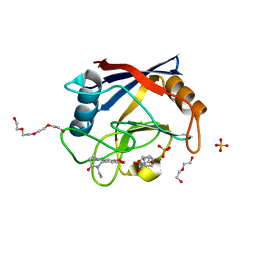 | | Human Cyclophilin D in complex with norbornane fragment derivative | | Descriptor: | 14-ethyl-4,6-dioxa-10,14-diazatricyclo[7.6.0.0^{3,7}]pentadeca-1(9),2,7-trien-13-one, 2-[(1~{R},2~{R},6~{S},7~{S})-3,5-bis(oxidanylidene)-4-azatricyclo[5.2.1.0^{2,6}]decan-4-yl]ethanoic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Graedler, U. | | Deposit date: | 2019-04-05 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel Cyclophilin D inhibitors starting from three dimensional fragments with millimolar potencies.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4UCC
 
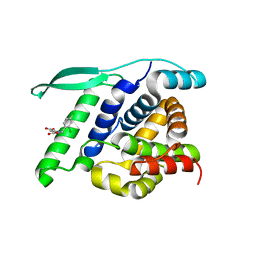 | | N-terminal globular domain of the RSV Nucleoprotein in complex with the Nucleoprotein Phosphoprotein interaction inhibitor M76 | | Descriptor: | 1-[(2,4-dichlorophenyl)methyl]pyrazole-3,5-dicarboxylic acid, NUCLEOPROTEIN | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
