5ONS
 
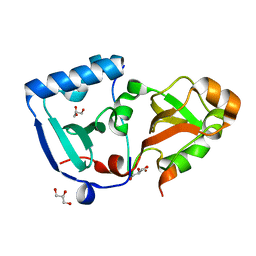 | | Crystal structure of the minimal DENR-MCTS1 complex | | Descriptor: | Density-regulated protein, GLYCEROL, Malignant T-cell-amplified sequence 1, ... | | Authors: | Ahmed, Y.L, Sinning, I. | | Deposit date: | 2017-08-04 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | DENR-MCTS1 heterodimerization and tRNA recruitment are required for translation reinitiation.
PLoS Biol., 16, 2018
|
|
3OIJ
 
 | |
3J0O
 
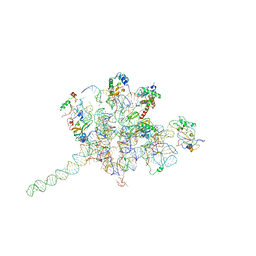 | | Core of mammalian 80S pre-ribosome in complex with tRNAs fitted to a 9A cryo-EM map: classic PRE state 2 | | Descriptor: | 40S ribosomal RNA fragment, 60S ribosomal RNA fragment, Ribosomal protein L10a, ... | | Authors: | Budkevich, T, Giesebrecht, J, Altman, R, Munro, J, Mielke, T, Nierhaus, K, Blanchard, S, Spahn, C.M. | | Deposit date: | 2011-10-05 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure and dynamics of the Mammalian ribosomal pretranslocation complex.
Mol.Cell, 44, 2011
|
|
3J0P
 
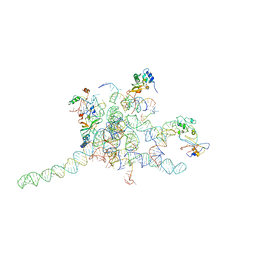 | | Core of mammalian 80S pre-ribosome in complex with tRNAs fitted to a 10.6A cryo-em map: rotated PRE state 1 | | Descriptor: | 40S ribosomal RNA fragment, 60S ribosomal RNA fragment, Ribosomal protein L10a, ... | | Authors: | Budkevich, T, Giesebrecht, J, Altman, R, Munro, J, Mielke, T, Nierhaus, K, Blanchard, S, Spahn, C.M. | | Deposit date: | 2011-10-06 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.6 Å) | | Cite: | Structure and dynamics of the Mammalian ribosomal pretranslocation complex.
Mol.Cell, 44, 2011
|
|
3P87
 
 | | Structure of human PCNA bound to RNASEH2B PIP box peptide | | Descriptor: | Proliferating cell nuclear antigen, Ribonuclease H2 subunit B | | Authors: | Bubeck, D, Reijns, M.A, Graham, S.C, Astell, K.R, Jones, E.Y, Jackson, A.P. | | Deposit date: | 2010-10-13 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | PCNA directs type 2 RNase H activity on DNA replication and repair substrates.
Nucleic Acids Res., 39, 2011
|
|
2P40
 
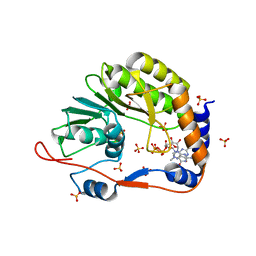 | |
5L7P
 
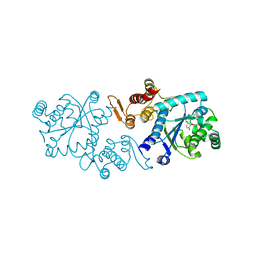 | | In silico-powered specific incorporation of photocaged Dopa at multiple protein sites | | Descriptor: | (2~{S})-2-azanyl-3-[3-[(2-nitrophenyl)methoxy]-4-oxidanyl-phenyl]propanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hauf, M, Richter, F, Schneider, T, Martins, B.M, Baumann, T, Durkin, P, Dobbek, H, Moeglich, A, Budisa, N. | | Deposit date: | 2016-06-03 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Photoactivatable Mussel-Based Underwater Adhesive Proteins by an Expanded Genetic Code.
Chembiochem, 18, 2017
|
|
4AIM
 
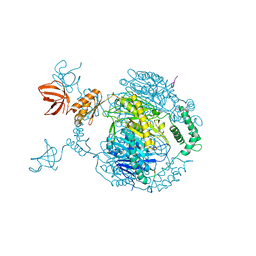 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | Descriptor: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | Authors: | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | Deposit date: | 2012-02-10 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
4AID
 
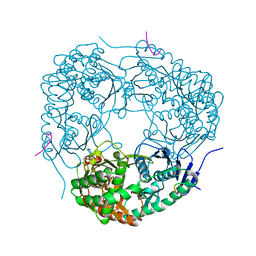 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | Descriptor: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | Authors: | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | Deposit date: | 2012-02-09 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
3V9W
 
 | |
3CV7
 
 | | Crystal structure of porcine aldehyde reductase ternary complex | | Descriptor: | 3,5-dichloro-2-hydroxybenzoic acid, Alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2008-04-18 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Structure of aldehyde reductase in ternary complex with coenzyme and the potent 20alpha-hydroxysteroid dehydrogenase inhibitor 3,5-dichlorosalicylic acid: Implications for inhibitor binding and selectivity
Arch.Biochem.Biophys., 479, 2008
|
|
4M57
 
 | | Crystal structure of the pentatricopeptide repeat protein PPR10 from maize | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10 | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
3V9U
 
 | |
3VA3
 
 | |
3DAT
 
 | |
3V9X
 
 | |
3VA0
 
 | |
3DAU
 
 | |
1XBP
 
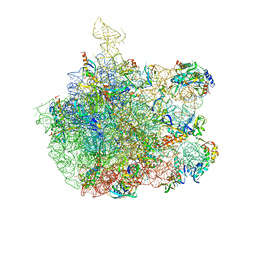 | | Inhibition of peptide bond formation by pleuromutilins: The structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with Tiamulin | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Schluenzen, F, Pyetan, E, Fucini, P, Yonath, A, Harms, J.M. | | Deposit date: | 2004-08-31 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of peptide bond formation by pleuromutilins: the structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin.
Mol.Microbiol., 54, 2004
|
|
6EXN
 
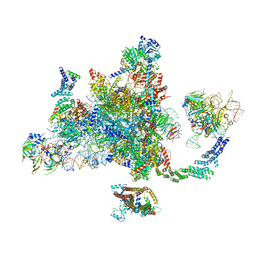 | | Post-catalytic P complex spliceosome with 3' splice site docked | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat: UBC4 RNA, ... | | Authors: | Wilkinson, M.E, Fica, S.M, Galej, W.P, Norman, C.M, Newman, A.J, Nagai, K. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Postcatalytic spliceosome structure reveals mechanism of 3'-splice site selection.
Science, 358, 2017
|
|
2CQH
 
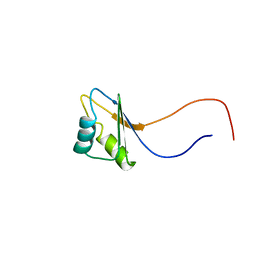 | | Solution structure of the RNA binding domain of IGF-II mRNA-binding protein 2 | | Descriptor: | IGF-II mRNA-binding protein 2 isoform a | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain of IGF-II mRNA-binding protein 2
To be Published
|
|
7N0B
 
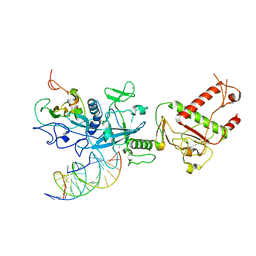 | | Cryo-EM structure of SARS-CoV-2 nsp10-nsp14 (WT)-RNA complex | | Descriptor: | CALCIUM ION, Non-structural protein 10, Proofreading exoribonuclease, ... | | Authors: | Liu, C, Yang, Y. | | Deposit date: | 2021-05-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme.
Science, 373, 2021
|
|
7N0D
 
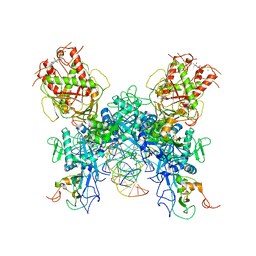 | |
7N0C
 
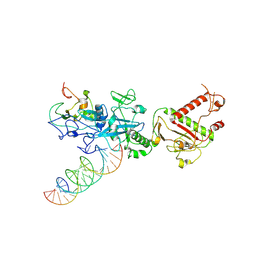 | |
7BMK
 
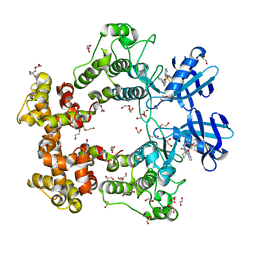 | | ATP-Competitive Partial Antagonists-'PAIR's-Rheostatically Modulate IRE1alpha's Kinase Helix-alphaC to Segregate its RNase-Mediated Biological Outputs | | Descriptor: | 1,2-ETHANEDIOL, 2,2,2-tris(fluoranyl)-~{N}-[4-[3-[2-[[(3~{S})-piperidin-3-yl]amino]pyrimidin-4-yl]pyridin-2-yl]oxynaphthalen-1-yl]ethanesulfonamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feldman, H.C, Ghosh, R, Auyeung, V, Mueller, J.L, Vidadala, V.N, Olivier, A, Backes, B.J, Zikherman, J, Papa, F.R, Maly, D.J. | | Deposit date: | 2021-01-20 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | ATP-competitive partial antagonists of the IRE1 alpha RNase segregate outputs of the UPR.
Nat.Chem.Biol., 17, 2021
|
|
