3MV0
 
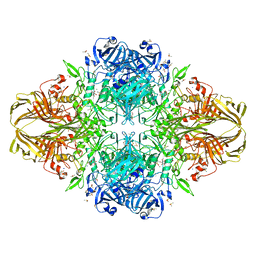 | | E. COLI (lacZ) BETA-GALACTOSIDASE (R599A) IN COMPLEX WITH D-GALCTOPYRANOSYL-1-ONE | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Vance, M, Driedger, M.L, Nibber, A, Tran, A, Huber, R.E. | | Deposit date: | 2010-05-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Importance of Arg-599 of b-galactosidase (Escherichia coli) as an anchor for the open conformations of Phe-601 and the active-site loop
Biochem.Cell Biol., 88, 2010
|
|
7NBU
 
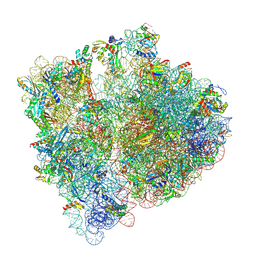 | | Structure of the HigB1 toxin mutant K95A from Mycobacterium tuberculosis (Rv1955) and its target, the cspA mRNA, on the E. coli Ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Giudice, E, Mansour, M, Chat, S, D'Urso, G, Gillet, R, Genevaux, P. | | Deposit date: | 2021-01-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Substrate recognition and cryo-EM structure of the ribosome-bound TAC toxin of Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
6ELI
 
 | | Structure of HIV-1 reverse transcriptase (RT) in complex with rilpivirine and an RNase H inhibitor XZ462 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Gag-Pol polyprotein, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2017-09-29 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Developing and Evaluating Inhibitors against the RNase H Active Site of HIV-1 Reverse Transcriptase.
J. Virol., 92, 2018
|
|
5TL9
 
 | | crystal structure of mPGES-1 bound to inhibitor | | Descriptor: | 2-{2-[(1S,2S)-2-{[1-(8-methylquinolin-2-yl)piperidine-4-carbonyl]amino}cyclopentyl]ethyl}benzoic acid, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Luz, J.G, Antonysamy, S, Partridge, K, Fisher, M. | | Deposit date: | 2016-10-10 | | Release date: | 2017-03-01 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery and characterization of [(cyclopentyl)ethyl]benzoic acid inhibitors of microsomal prostaglandin E synthase-1.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
3H1C
 
 | |
3MUZ
 
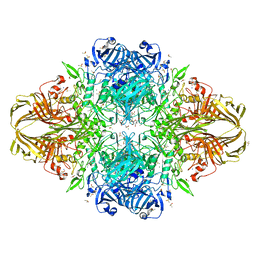 | | E.Coli (lacZ) beta-galactosidase (R599A) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Vance, M.L, Driedger, M.R, Nibber, A, Tran, A, Huber, R.E. | | Deposit date: | 2010-05-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Importance of Arg-599 of b-galactosidase (Escherichia coli) as an anchor for the open conformations of Phe-601 and the active-site loop
Biochem.Cell Biol., 88, 2010
|
|
1K4R
 
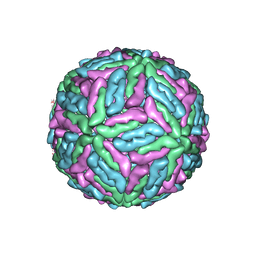 | | Structure of Dengue Virus | | Descriptor: | MAJOR ENVELOPE PROTEIN E | | Authors: | Kuhn, R.J, Zhang, W, Rossmann, M.G, Pletnev, S.V, Corver, J, Lenches, E, Jones, C.T, Mukhopadhyay, S, Chipman, P.R, Strauss, E.G, Baker, T.S, Strauss, J.H. | | Deposit date: | 2001-10-08 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Structure of dengue virus: implications for flavivirus organization, maturation, and fusion.
Cell(Cambridge,Mass.), 108, 2002
|
|
3H8A
 
 | |
6L0R
 
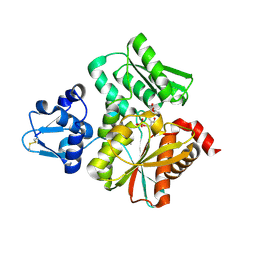 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Acetylserine | | Descriptor: | (2S)-3-acetyloxy-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
1HTL
 
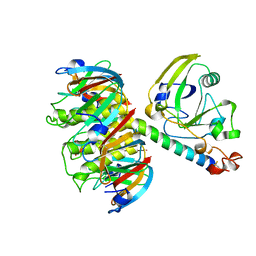 | |
1S1U
 
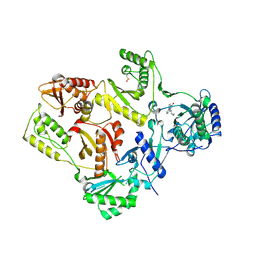 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1Y6V
 
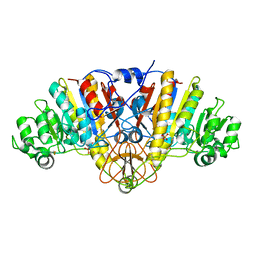 | | Structure of E. coli Alkaline Phosphatase in presence of cobalt at 1.60 A resolution | | Descriptor: | Alkaline phosphatase, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Wang, J, Stieglitz, K, Kantrowitz, E.R. | | Deposit date: | 2004-12-07 | | Release date: | 2005-06-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal Specificity Is Correlated with Two Crucial Active Site Residues in Escherichia coli Alkaline Phosphatase(,).
Biochemistry, 44, 2005
|
|
1Y55
 
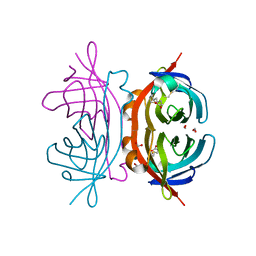 | | Crystal structure of the C122S mutant of E. Coli expressed avidin related protein 4 (AVR4)-biotin complex | | Descriptor: | Avidin-related protein 4/5, BIOTIN, FORMIC ACID | | Authors: | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Y7A
 
 | | Structure of D153H/K328W E. coli alkaline phosphatase in presence of cobalt at 1.77 A resolution | | Descriptor: | Alkaline phosphatase, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Wang, J, Stieglitz, K, Kantrowitz, E.R. | | Deposit date: | 2004-12-08 | | Release date: | 2005-06-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Metal Specificity Is Correlated with Two Crucial Active Site Residues in Escherichia coli Alkaline Phosphatase(,).
Biochemistry, 44, 2005
|
|
1KPM
 
 | | First Structural Evidence of a Specific Inhibition of Phospholipase A2 by Vitamin E and its Implications in Inflammation: Crystal Structure of the Complex Formed between Phospholipase A2 and Vitamin E at 1.8 A Resolution. | | Descriptor: | ACETIC ACID, Phospholipase A2, VITAMIN E | | Authors: | Chandra, V, Jasti, J, Kaur, P, Betzel, C, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-01-01 | | Release date: | 2002-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | First Structural Evidence of a Specific Inhibition of Phospholipase A2 by alpha-Tocopherol (Vitamin E) and its
Implications in Inflammation: Crystal Structure of the Complex Formed Between Phospholipase A2 and
alpha-Tocopherol at 1.8 A Resolution
J.Mol.Biol., 320, 2002
|
|
1Z66
 
 | | NMR solution structure of domain III of E-protein of tick-borne Langat flavivirus (no RDC restraints) | | Descriptor: | Major envelope protein E | | Authors: | Mukherjee, M, Dutta, K, White, M.A, Cowburn, D, Fox, R.O. | | Deposit date: | 2005-03-21 | | Release date: | 2006-03-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and backbone dynamics of domain III of the E protein of tick-borne Langat flavivirus suggests a potential site for molecular recognition.
Protein Sci., 15, 2006
|
|
7BH1
 
 | | Cryo-EM Structure of KdpFABC in E1 state with K | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BGY
 
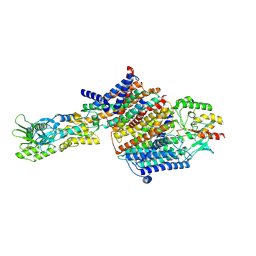 | | Cryo-EM Structure of KdpFABC in E2Pi state with MgF4 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, MAGNESIUM ION, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KA1
 
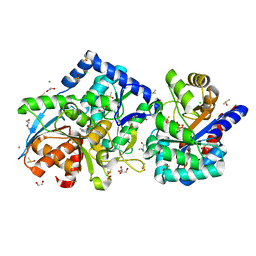 | | 1.60 Angstrom resolution crystal structure of the beta-Q114A mutant Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and cesium ion at the metal coordination site | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.60 Angstrom resolution crystal structure of the beta-Q114A mutant Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and cesium ion at the metal coordination site.
To be Published
|
|
4LQ3
 
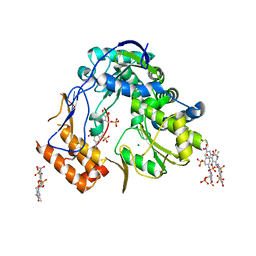 | | Crystal structure of human norovirus RNA-dependent RNA-polymerase bound to the inhibitor PPNDS | | Descriptor: | 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, 5'-R(P*GP*G)-3', MAGNESIUM ION, ... | | Authors: | Milani, M, Tarantino, D, Mastrangelo, E, Croci, R. | | Deposit date: | 2013-07-17 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Naphthalene-sulfonate inhibitors of human norovirus RNA-dependent RNA-polymerase.
Antiviral Res., 102, 2014
|
|
7LC3
 
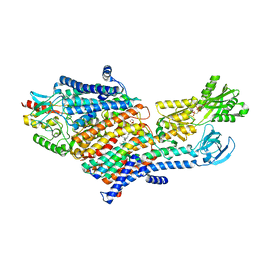 | | CryoEM Structure of KdpFABC in E1-ATP state | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BNL
 
 | | Notum ARUK3003710 | | Descriptor: | (4~{E})-2-(3,4-dimethylphenyl)-4-[(1-methylpyrazol-4-yl)methylidene]-1,3-oxazol-5-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P.V, Svensson, F, Steadman, D. | | Deposit date: | 2021-01-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Notum Inhibitor ARUK3003710
To Be Published
|
|
7KXC
 
 | | The aminoacrylate form of the wild-type Salmonella typhimurium Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, sodium ion at the metal coordination site and benzimidazole (BZI) at the enzyme beta-site at 1.30 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-12-03 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The aminoacrylate form of the wild-type Salmonella typhimurium Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, sodium ion at the metal coordination site and benzimidazole (BZI) at the enzyme beta-site at 1.30 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring.
To be Published
|
|
7KWV
 
 | |
