8ZMU
 
 | |
9EO4
 
 | | Outward-open structure of human dopamine transporter bound to cocaine | | Descriptor: | CHLORIDE ION, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Nielsen, J.C, Salomon, K, Kalenderoglou, I.E, Bargmeyer, S, Pape, T, Shahsavar, A, Loland, C.J. | | Deposit date: | 2024-03-14 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | The molecular basis for cocaine binding to the human dopamine transporter
To Be Published
|
|
5VEZ
 
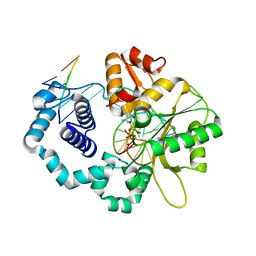 | | DNA polymerase beta substrate complex with 8-oxoG:A at the primer terminus and incoming dCTP analog | | Descriptor: | 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (5'-D(*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), ... | | Authors: | Freudenthal, B.D, Schaich, M.A, Whitaker, A.M, Smith, M.R. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Capturing a mammalian DNA polymerase extending from an oxidized nucleotide.
Nucleic Acids Res., 45, 2017
|
|
5W2Y
 
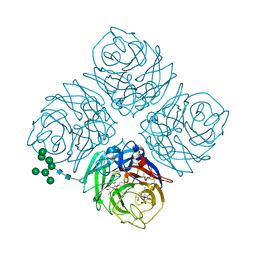 | | INFLUENZA VIRUS NEURAMINIDASE N9 IN COMPLEX WITH 9-DEOXYGENATED 2,3-DIFLUORO-N-ACETYLNEURAMINIC ACID | | Descriptor: | (2R,3R,4R,5R,6R)-5-acetamido-6-[(1R,2R)-1,2-bis(oxidanyl)propyl]-2,3-bis(fluoranyl)-4-oxidanyl-oxane-2-carboxylic acid, (2~{R},3~{R},4~{R},5~{R})-3-acetamido-2-[(1~{R},2~{R})-1,2-bis(oxidanyl)propyl]-5-fluoranyl-4-oxidanyl-2,3,4,5-tetrahydropyran-1-ium-6-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Mckimm-Breschkin, J, Barrett, S, Pilling, P, Hader, S, Watt, A.G. | | Deposit date: | 2017-06-07 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and Functional Analysis of Anti-Influenza Activity of 4-, 7-, 8- and 9-Deoxygenated 2,3-Difluoro- N-acetylneuraminic Acid Derivatives.
J. Med. Chem., 61, 2018
|
|
5DDR
 
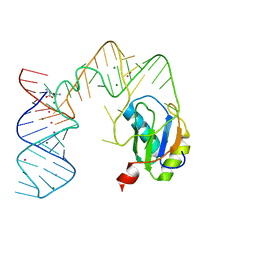 | | L-glutamine riboswitch bound with L-glutamine soaked with Cs+ | | Descriptor: | CESIUM ION, GLUTAMINE, L-glutamine riboswitch RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
6JY3
 
 | | Monomeric Form of Bovine Heart Cytochrome c Oxidase in the Fully Oxidized State | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (2S,3S,4S,5S,6R)-2-(2-decoxyethoxy)-6-(hydroxymethyl)oxane-3,4,5-triol, ... | | Authors: | Shinzawa-Itoh, K, Muramoto, K. | | Deposit date: | 2019-04-26 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Monomeric structure of an active form of bovine cytochromecoxidase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2XCD
 
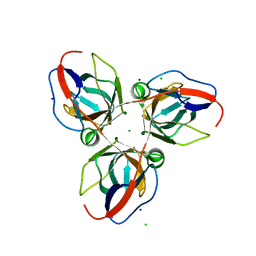 | | Structure of YncF,the genomic dUTPase from Bacillus subtilis | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Garcia, J, Burchell, L, Takezawa, M, Rzechorzek, N.J, Fogg, M, Wilson, K.S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Structure of the Genomic Bacillus Subtilis Dutpase: Novel Features in the Phe-Lid.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6D7A
 
 | | Structure of T. gondii PLP1 beta-rich domain | | Descriptor: | Perforin-like protein 1, SODIUM ION | | Authors: | Guerra, A.J, Koropatkin, N.M, Wawrzak, Z, Bahr, C.M.E, Carruthers, V.B. | | Deposit date: | 2018-04-24 | | Release date: | 2018-05-16 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structural basis of Toxoplasma gondii perforin-like protein 1 membrane interaction and activity during egress.
PLoS Pathog., 14, 2018
|
|
6ZE8
 
 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound OATD-01 | | Descriptor: | 5-(4-((2S,5S)-5-(4-chlorobenzyl)-2-methylmorpholino)piperidin-1-yl)-4H-1,2,4-triazol-3-amine, Chitotriosidase-1, GLYCEROL, ... | | Authors: | Nowotny, M, Bartlomiejczak, A, Napiorkowska-Gromadzka, A. | | Deposit date: | 2020-06-16 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of OATD-01 , a First-in-Class Chitinase Inhibitor as Potential New Therapeutics for Idiopathic Pulmonary Fibrosis.
J.Med.Chem., 63, 2020
|
|
6DJQ
 
 | | Vps1 GTPase-BSE fusion complexed with GDP.AlF4- | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Varlakhanova, N.V, Brady, T.M, Tornabene, B.A, Hosford, C.J, Chappie, J.S, Ford, M.G.J. | | Deposit date: | 2018-05-25 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
2XBY
 
 | | Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor | | Descriptor: | (3R,4R)-1-METHYLCARBAMOYLMETHYL-PYRROLIDINE-3,4-DICARBOXYLIC ACID 3-[(4-CHLORO-PHENYL)-AMIDE] 4-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYL]-AMIDE}, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Banner, D.W, Benz, J, Schlatter, D, Thomi, S, Haap, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery of a Factor Xa Inhibitor (3R,4R)-1-(2,2-Difluoro-Ethyl)-Pyrrolidine-3,4-Dicarboxylic Acid 3-[(5-Chloro-Pyridin-2-Yl)-Amide] 4-{[2-Fluoro-4-(2-Oxo-2H-Pyridin-1-Yl)-Phenyl]-Amide} as a Clinical Candidate.
Bioorg.Med.Chem., 20, 2010
|
|
2XBX
 
 | | Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor | | Descriptor: | (3R,4R)-1-METHANESULFONYL-PYRROLIDINE-3,4-DICARBOXYLIC ACID 3-[(4-CHLORO-PHENYL)-AMIDE] 4-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYL]-AMIDE}, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Banner, D.W, Benz, J, Schlatter, D, Haap, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of a Factor Xa Inhibitor (3R,4R)-1-(2,2-Difluoro-Ethyl)-Pyrrolidine-3,4-Dicarboxylic Acid 3-[(5-Chloro-Pyridin-2-Yl)-Amide] 4-{[2-Fluoro-4-(2-Oxo-2H-Pyridin-1-Yl)-Phenyl]-Amide} as a Clinical Candidate.
Bioorg.Med.Chem., 20, 2010
|
|
2XBW
 
 | | Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor | | Descriptor: | (3R,4R)-1-SULFAMOYL-PYRROLIDINE-3,4-DICARBOXYLIC ACID 3-[(4-CHLORO-PHENYL)-AMIDE] 4-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYL]-AMIDE}, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Banner, D.W, Benz, J, Schlatter, D, Thomi, S, Haap, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of a Factor Xa Inhibitor (3R,4R)-1-(2,2-Difluoro-Ethyl)-Pyrrolidine-3,4-Dicarboxylic Acid 3-[(5-Chloro-Pyridin-2-Yl)-Amide] 4-{[2-Fluoro-4-(2-Oxo-2H-Pyridin-1-Yl)-Phenyl]-Amide} as a Clinical Candidate.
Bioorg.Med.Chem., 20, 2010
|
|
6QQ3
 
 | | The room temperature structure of lysozyme via the acoustic levitation of a droplet | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Axford, D.N, Docker, P, Dye, E, Morris, R. | | Deposit date: | 2019-02-17 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Non-Contact Universal Sample Presentation for Room Temperature Macromolecular Crystallography Using Acoustic Levitation.
Sci Rep, 9, 2019
|
|
6QXA
 
 | | Structure of membrane bound pyrophosphatase from Thermotoga maritima in complex with imidodiphosphate and N-[(2-amino-6-benzothiazolyl)methyl]-1H-indole-2-carboxamide (ATC) | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, IMIDODIPHOSPHORIC ACID, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Vidilaseris, K, Goldman, A. | | Deposit date: | 2019-03-07 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Asymmetry in catalysis byThermotoga maritimamembrane-bound pyrophosphatase demonstrated by a nonphosphorus allosteric inhibitor.
Sci Adv, 5, 2019
|
|
6KW1
 
 | | The structure of the metallo-beta-lactamase VIM-2 in complex with a triazolylthioacetamide 1b | | Descriptor: | 2-[3-[2-(1H-benzimidazol-2-ylamino)-2-oxidanylidene-ethyl]sulfanyl-1H-1,2,4-triazol-5-yl]benzoic acid, ACETATE ION, Beta-lactamase class B VIM-2, ... | | Authors: | Yang, K.W, Xiang, Y. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.775212 Å) | | Cite: | Kinetic, Thermodynamic, and Crystallographic Studies of 2-Triazolylthioacetamides as Verona Integron-Encoded Metallo-beta-Lactamase 2 (VIM-2) Inhibitor.
Biomolecules, 10, 2020
|
|
5KH9
 
 | | Crystal structure of a low occupancy fragment candidate (5-[(4-Isopropylphenyl)amino]-6-methyl-1,2,4-triazin-3(2H)-one) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 6-methyl-5-[(4-propan-2-ylphenyl)amino]-2~{H}-1,2,4-triazin-3-one, FORMIC ACID, Histone deacetylase 6, ... | | Authors: | Harding, R.J, Tempel, W, Ravichandran, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, Schapira, M, Bountra, C, Edwards, A.M, von Delft, F, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
4W5Z
 
 | |
3HIU
 
 | | The crystal structure of protein (XCC3681) from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Uncharacterized protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-20 | | Release date: | 2009-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of protein (XCC3681) from Xanthomonas campestris pv. campestris str. ATCC 33913
To be Published
|
|
3HIJ
 
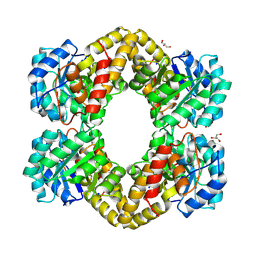 | | Crystal structure of dihydrodipicolinate synthase from Bacillus anthracis in complex with its substrate, pyruvate | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, SODIUM ION | | Authors: | Voss, J.E, Scally, S.W, Dobson, R.C.J, Perugini, M.A. | | Deposit date: | 2009-05-20 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Substrate-mediated Stabilization of a Tetrameric Drug Target Reveals Achilles Heel in Anthrax.
J.Biol.Chem., 285, 2010
|
|
3HJB
 
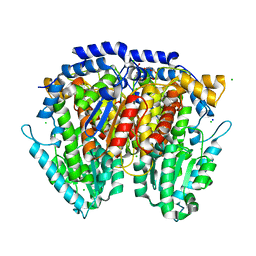 | | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae.
To be Published
|
|
4F5N
 
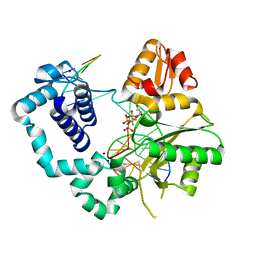 | | Open ternary complex of R283K DNA polymerase beta with a metal free dCTP analog | | Descriptor: | 2'-deoxy-5'-O-[(S)-{difluoro[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}(hydroxy)phosphoryl]cytidine, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Beard, W.A, Wilson, S.H. | | Deposit date: | 2012-05-13 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of dNTP Intermediate States during DNA Polymerase Active Site Assembly.
Structure, 20, 2012
|
|
4F3Y
 
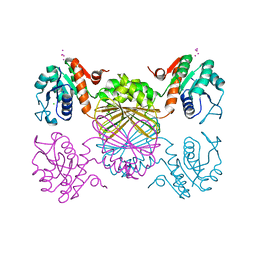 | |
6Z74
 
 | |
6QDT
 
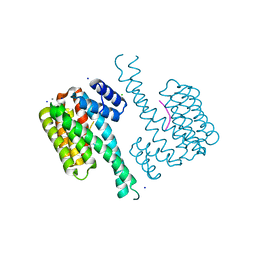 | | Crystal structure of 14-3-3sigma in complex with a RapGef2 pT740 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Kaplan, A, Fournier, A.E, Ottman, C. | | Deposit date: | 2019-01-02 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Polypharmacological Perturbation of the 14-3-3 Adaptor Protein Interactome Stimulates Neurite Outgrowth.
Cell Chem Biol, 27, 2020
|
|
