2OJI
 
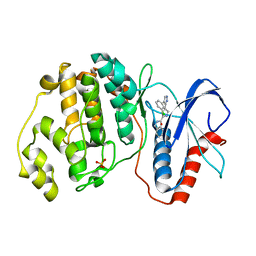 | |
1FV5
 
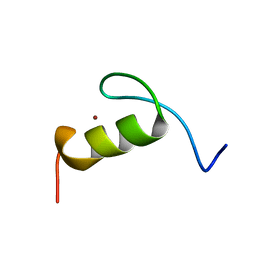 | | SOLUTION STRUCTURE OF THE FIRST ZINC FINGER FROM THE DROSOPHILA U-SHAPED TRANSCRIPTION FACTOR | | Descriptor: | FIRST ZINC FINGER OF U-SHAPED, ZINC ION | | Authors: | Liew, C.K, Kowalski, K, Fox, A.H, Newton, A, Sharpe, B.K, Crossley, M, Mackay, J.P. | | Deposit date: | 2000-09-18 | | Release date: | 2000-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two CCHC zinc fingers from the FOG family protein U-shaped that mediate protein-protein interactions.
Structure Fold.Des., 8, 2000
|
|
2OJJ
 
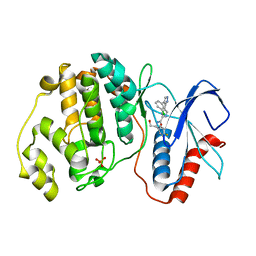 | |
8VEW
 
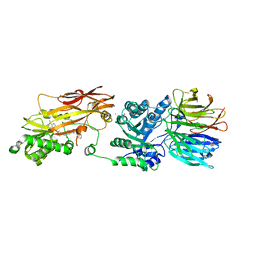 | | Crystal structure of PRMT5:MEP50 in complex with MTA and oxamide compound 24 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5-{2-[(2R,5S)-5-methyl-2-phenylpiperidin-1-yl](oxo)acetamido}pyridine-3-carboxamide, CHLORIDE ION, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
8VEO
 
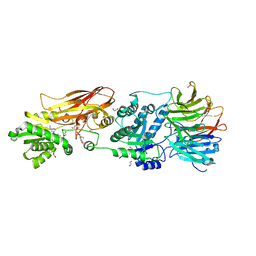 | | Crystal structure of PRMT5:MEP50 in complex with MTA | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
5ZHX
 
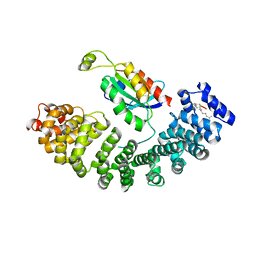 | |
2OJG
 
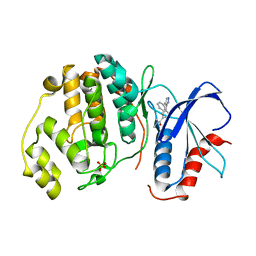 | |
1BX6
 
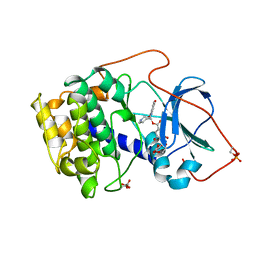 | | CRYSTAL STRUCTURE OF THE POTENT NATURAL PRODUCT INHIBITOR BALANOL IN COMPLEX WITH THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE | | Descriptor: | BALANOL, CAMP-DEPENDENT PROTEIN KINASE | | Authors: | Narayana, N, Xuong, N.-H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 1998-10-13 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the potent natural product inhibitor balanol in complex with the catalytic subunit of cAMP-dependent protein kinase.
Biochemistry, 38, 1999
|
|
1BKX
 
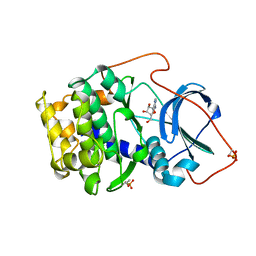 | | A BINARY COMPLEX OF THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE AND ADENOSINE FURTHER DEFINES CONFORMATIONAL FLEXIBILITY | | Descriptor: | ADENOSINE MONOPHOSPHATE, CAMP-DEPENDENT PROTEIN KINASE | | Authors: | Narayana, N, Cox, S, Xuong, N, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 1997-07-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A binary complex of the catalytic subunit of cAMP-dependent protein kinase and adenosine further defines conformational flexibility.
Structure, 5, 1997
|
|
3UVN
 
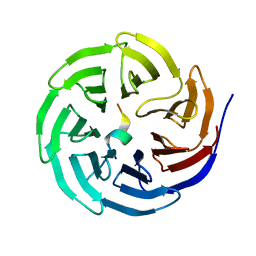 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of SET1A | | Descriptor: | Histone-lysine N-methyltransferase SETD1A, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
2WZR
 
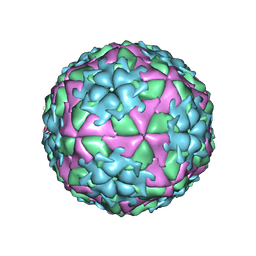 | | The Structure of Foot and Mouth Disease Virus Serotype SAT1 | | Descriptor: | POLYPROTEIN | | Authors: | Adams, P, Lea, S, Newman, J, Blakemore, W, King, A, Stuart, D, Fry, E. | | Deposit date: | 2009-12-02 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Foot-and-Mouth Disease Virus Serotype Sat1.
To be Published
|
|
8DY5
 
 | |
8DY1
 
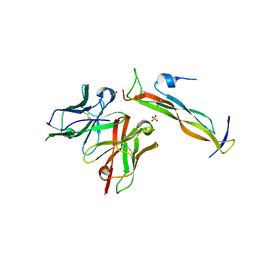 | |
1FU9
 
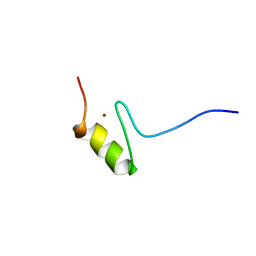 | | SOLUTION STRUCTURE OF THE NINTH ZINC-FINGER DOMAIN OF THE U-SHAPED TRANSCRIPTION FACTOR | | Descriptor: | U-SHAPED TRANSCRIPTIONAL COFACTOR, ZINC ION | | Authors: | Liew, C.K, Kowalski, K, Fox, A.H, Newton, A, Sharpe, B.K, Crossley, M, Mackay, J.P. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two CCHC zinc fingers from the FOG family protein U-shaped that mediate protein-protein interactions.
Structure Fold.Des., 8, 2000
|
|
3MSX
 
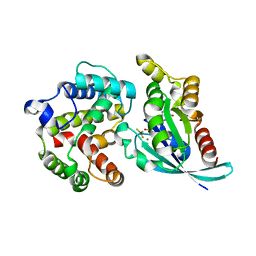 | | Crystal structure of RhoA.GDP.MgF3 in complex with GAP domain of ArhGAP20 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rho GTPase-activating protein 20, ... | | Authors: | Utepbergenov, D, Cooper, D.R, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2010-04-29 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of molecular specificity of RhoGAP domains towards small GTPases of RhoA family.
To be Published
|
|
5WFC
 
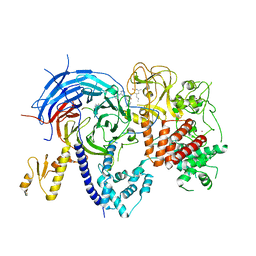 | |
1AUI
 
 | | HUMAN CALCINEURIN HETERODIMER | | Descriptor: | CALCIUM ION, FE (III) ION, SERINE/THREONINE PHOSPHATASE 2B, ... | | Authors: | Kissinger, C.R, Parge, H.E, Knighton, D.R, Pelletier, L.A, Lewis, C.T, Tempczyk, A, Villafranca, J.E. | | Deposit date: | 1997-08-27 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex.
Nature, 378, 1995
|
|
3UR4
 
 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Dong, A, Dombrovski, L, Senisterra, G, Wernimont, A, Wasney, G.A, Allali Hassani, A, Nguyen, K.T, Smil, D, Bolshan, Y, Hajian, T, Poda, G, Chau, I, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Brown, P, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-21 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
3UV4
 
 | | Crystal Structure of the second bromodomain of human Transcription initiation factor TFIID subunit 1 (TAF1) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3UVL
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of MLL3 | | Descriptor: | Histone-lysine N-methyltransferase MLL3, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
6IN9
 
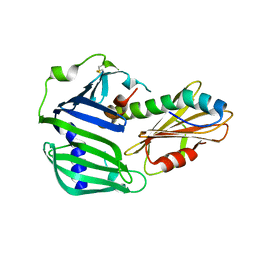 | | Crystal structure of MucB in complex with MucA(peri) | | Descriptor: | Sigma factor AlgU negative regulatory protein, Sigma factor AlgU regulatory protein MucB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
3QZT
 
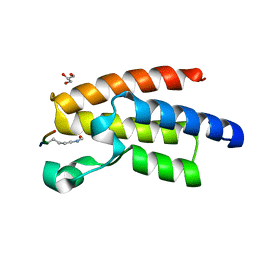 | | Crystal Structure of BPTF bromo in complex with histone H4K16ac - Form II | | Descriptor: | GLYCEROL, Histone H4, Nucleosome-remodeling factor subunit BPTF | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
3UVM
 
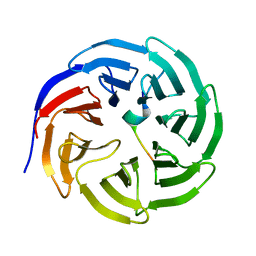 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of MLL4 | | Descriptor: | Histone-lysine N-methyltransferase MLL4, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
1D2N
 
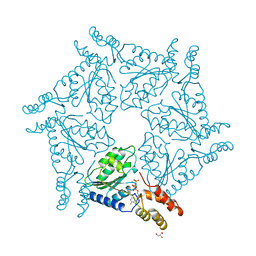 | | D2 DOMAIN OF N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN, ... | | Authors: | Lenzen, C.U, Steinmann, D, Whiteheart, S.W, Weis, W.I. | | Deposit date: | 1998-06-30 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein.
Cell(Cambridge,Mass.), 94, 1998
|
|
3UV5
 
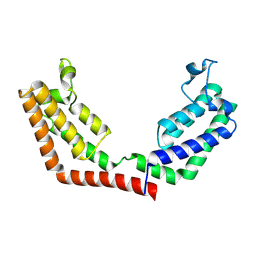 | | Crystal Structure of the tandem bromodomains of human Transcription initiation factor TFIID subunit 1 (TAF1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcription initiation factor TFIID subunit 1 | | Authors: | Filippakopoulos, P, Felletar, I, Picaud, S, Keates, T, Muniz, J, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-29 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
