6IBV
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with broad spectrum boronic inhibitor cpd 1 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Celenza, G, Venturelli, A, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6Q35
 
 | | Crystal structure of GES-5 beta-lactamase in complex with boronic inhibitor cpd 3 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, DIMETHYL SULFOXIDE, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
7R9L
 
 | | Crystal structure of HPK1 in complex with compound 2 | | Descriptor: | 2-amino-N,N-dimethyl-5-(1H-pyrrolo[2,3-b]pyridin-5-yl)benzamide, Hematopoietic progenitor kinase | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
7R9P
 
 | | Crystal structure of HPK1 in complex with compound 14 | | Descriptor: | 6-amino-2-fluoro-N,N-dimethyl-3-(4'-methylspiro[cyclopropane-1,3'-pyrrolo[2,3-b]pyridin]-5'-yl)benzamide, Hematopoietic progenitor kinase, SULFATE ION | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
7R9N
 
 | | Crystal structure of HPK1 in complex with GNE1858 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hematopoietic progenitor kinase, ... | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
6Q30
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with boronic inhibitor cpd 5 | | Descriptor: | (7-carboxy-1-benzothiophen-2-yl)-tris(oxidanyl)boranuide, CALCIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Celenza, G, Venturelli, A, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6ZAC
 
 | | PI3K Delta in complex with [(dimethylamino)methyldihydrobenzoxazin2methoxypyridinyl]methanesulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[5-[6-[(dimethylamino)methyl]-2,3-dihydro-1,4-benzoxazin-4-yl]-2-methoxy-pyridin-3-yl]methanesulfonamide | | Authors: | Convery, M.A, Hardy, C.J, Spencer, J.A, Rowland, P. | | Deposit date: | 2020-06-05 | | Release date: | 2020-07-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design and Development of a Macrocyclic Series Targeting Phosphoinositide 3-Kinase delta.
Acs Med.Chem.Lett., 11, 2020
|
|
6Q2Y
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with broad spectrum boronic inhibitor cpd3 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
7R53
 
 | | Crystal structure of human TLR8 in complex with Compound 15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-cyclopropyl-6-(2,6-dimethylpyridin-4-yl)-~{N}-[(3~{R},4~{R})-3-fluoranylpiperidin-4-yl]-1~{H}-indazol-3-amine, Toll-like receptor 8, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.121 Å) | | Cite: | Structure-Based Optimization of a Fragment-like TLR8 Binding Screening Hit to an In Vivo Efficacious TLR7/8 Antagonist.
Acs Med.Chem.Lett., 13, 2022
|
|
7R52
 
 | | Crystal structure of human TLR8 in complex with Compound 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methoxy-6-pyridin-4-yl-1~{H}-indole, Toll-like receptor 8, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.943 Å) | | Cite: | Structure-Based Optimization of a Fragment-like TLR8 Binding Screening Hit to an In Vivo Efficacious TLR7/8 Antagonist.
Acs Med.Chem.Lett., 13, 2022
|
|
7R54
 
 | | Crystal structure of human TLR8 in complex with Compound 4 | | Descriptor: | (5-methoxy-6-pyridin-4-yl-1~{H}-indazol-3-yl)-(4-methylpiperazin-1-yl)methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.836 Å) | | Cite: | Structure-Based Optimization of a Fragment-like TLR8 Binding Screening Hit to an In Vivo Efficacious TLR7/8 Antagonist.
Acs Med.Chem.Lett., 13, 2022
|
|
6YWL
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, ... | | Authors: | Schroeder, M, Ni, X, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
7RC9
 
 | | Crystal structure of human TLR8 ectodomain bound to small molecule antagonist 21 | | Descriptor: | 2-(2,6-dimethylpyridin-4-yl)-5-(piperidin-4-yl)-3-(propan-2-yl)-1H-indole, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Critton, D.A. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Identification of 2-Pyridinylindole-Based Dual Antagonists of Toll-like Receptors 7 and 8 (TLR7/8).
Acs Med.Chem.Lett., 13, 2022
|
|
7RSY
 
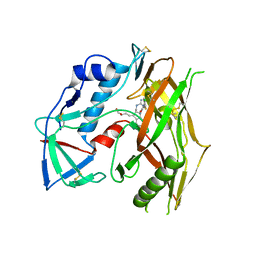 | | HIV-1 gp120 complex with CJF-III-049-R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 Clade C1086, N~1~-{(1R,2R,3S)-2-(carbamimidamidomethyl)-3-[(3R)-3,4-dihydroxybutyl]-5-[(methylamino)methyl]-2,3-dihydro-1H-inden-1-yl}-N~2~-(4-chloro-3-fluorophenyl)ethanediamide | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of gp120 Residue His105 as a Novel Target for HIV-1 Neutralization by Small-Molecule CD4-Mimics.
Acs Med.Chem.Lett., 12, 2021
|
|
7R01
 
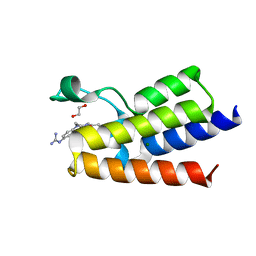 | | BAZ2A bromodomain in complex with N-acetylpiperazine derivative fragment 18 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(4-ethanoylpiperazin-1-yl)phenyl]guanidine, Bromodomain adjacent to zinc finger domain protein 2A, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.256 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
7R0B
 
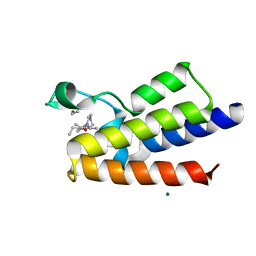 | | BAZ2A bromodomain in complex with acetylpyrrole derivative compound 47 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-ethyl-2-methyl-5-(2-piperazin-1-yl-1,3-thiazol-4-yl)-1~{H}-pyrrol-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2A, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
1TKB
 
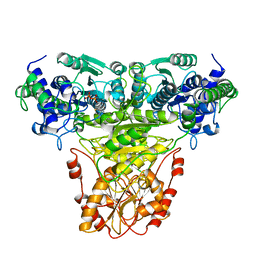 | |
1TKC
 
 | |
1TKA
 
 | |
6IBS
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with boronic inhibitor cpd 6 | | Descriptor: | CALCIUM ION, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6B3F
 
 | |
6XDM
 
 | |
6B3G
 
 | |
6MR5
 
 | |
6B36
 
 | |
