4NAO
 
 | | Crystal structure of EasH | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Janke, R, Havemann, J, Vogel, D, Keller, U, Loll, B. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Cyclolization of d-lysergic Acid alkaloid peptides.
Chem.Biol., 21, 2014
|
|
4NAP
 
 | | Crystal structure of a trap periplasmic solute binding protein from Desulfovibrio alaskensis G20 (DDE_0634), target EFI-510102, with bound d-tryptophan | | Descriptor: | D-TRYPTOPHAN, Extracellular solute-binding protein, family 7 | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NAQ
 
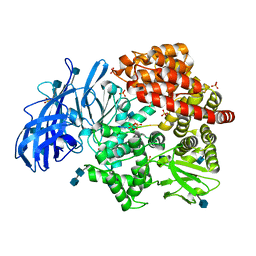 | | Crystal structure of porcine aminopeptidase-N complexed with poly-alanine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Chen, L, Lin, Y.L, Peng, G, Li, F. | | Deposit date: | 2013-10-22 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for multifunctional roles of mammalian aminopeptidase N.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4NAR
 
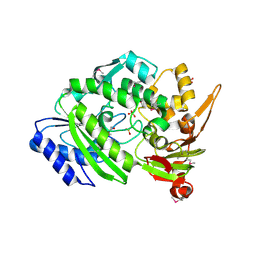 | | Crystal Structure of the Q9WYS3 protein from Thermotoga maritima. Northeast Structural Genomics Consortium Target VR152 | | Descriptor: | ACETATE ION, Putative uronate isomerase, SULFATE ION | | Authors: | Vorobiev, S, Lew, S, Seetharaman, J, Chi, Y, Xiao, R, Maglaqui, M, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Crystal Structure of the Q9WYS3 protein from Thermotoga maritima.
To be Published
|
|
4NAS
 
 | | The crystal structure of a rubisco-like protein (MtnW) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | CALCIUM ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-13 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The crystal structure of a rubisco-like protein (MtnW) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446.
To be Published
|
|
4NAT
 
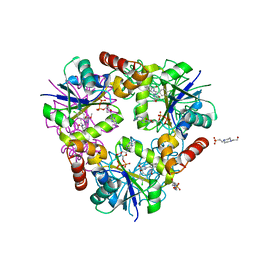 | | Inhibitors of 4-Phosphopanthetheine Adenylyltransferase | | Descriptor: | (1R,2R)-N-(3,4-dichlorobenzyl)-2-(4,6-dimethoxypyrimidin-2-yl)cyclohexanecarboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lahiri, S.D. | | Deposit date: | 2013-10-22 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Inhibitors of 4'-Phosphopantetheine Adenylyltransferase (PPAT) To Validate PPAT as a Target for Antibacterial Therapy.
Antimicrob.Agents Chemother., 57, 2013
|
|
4NAU
 
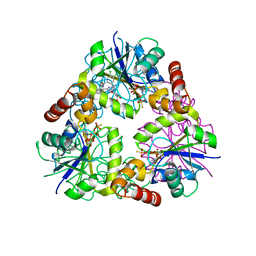 | | S. aureus CoaD with Inhibitor | | Descriptor: | 2-[2-[(1S,2S)-2-[(3,4-dichlorophenyl)methylcarbamoyl]cyclohexyl]-6-ethyl-pyrimidin-4-yl]-4-oxidanyl-6-oxidanylidene-1H-pyrimidine-5-carboxamide, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Phosphopantetheine adenylyltransferase | | Authors: | Lahiri, S.D. | | Deposit date: | 2013-10-22 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery of Inhibitors of 4'-Phosphopantetheine Adenylyltransferase (PPAT) To Validate PPAT as a Target for Antibacterial Therapy.
Antimicrob.Agents Chemother., 57, 2013
|
|
4NAV
 
 | | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608 | | Descriptor: | HYPOTHETICAL PROTEIN XCC279 | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608
TO BE PUBLISHED
|
|
4NAW
 
 | | Crystal Structure of Human ATG12~ATG5-ATG16N in complex with a fragment of ATG3 | | Descriptor: | Autophagy protein 5, Autophagy-related protein 16-1, SODIUM ION, ... | | Authors: | Metlagel, Z, Otomo, C, Takaesu, G, Otomo, T. | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Structural basis of ATG3 recognition by the autophagic ubiquitin-like protein ATG12.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4NAX
 
 | | Crystal structure of glutathione transferase PPUT_1760 from Pseudomonas putida, target EFI-507288, with one glutathione disulfide bound per one protein subunit | | Descriptor: | FORMIC ACID, GLYCEROL, Glutathione S-transferase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Crystal structure of glutathione transferase Pput_1760 from Pseudomonas putida, target EFI-507288
To be Published
|
|
4NAY
 
 | | Crystal Structure of FosB from Staphylococcus aureus with Zn and Sulfate at 1.42 Angstrom Resolution - SAD Phasing | | Descriptor: | Metallothiol transferase FosB, SULFATE ION, ZINC ION | | Authors: | Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
4NAZ
 
 | | Crystal Structure of FosB from Staphylococcus aureus with Zn and Sulfate at 1.15 Angstrom Resolution | | Descriptor: | GLYCEROL, Metallothiol transferase FosB, SULFATE ION, ... | | Authors: | Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
4NB0
 
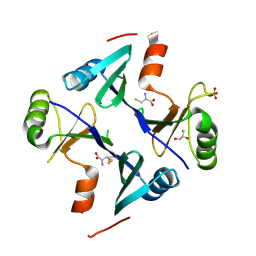 | | Crystal Structure of FosB from Staphylococcus aureus with BS-Cys9 disulfide at 1.62 Angstrom Resolution | | Descriptor: | CYSTEINE, GLYCEROL, Metallothiol transferase FosB, ... | | Authors: | Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
4NB1
 
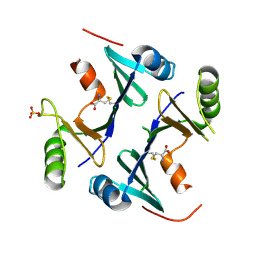 | | Crystal Structure of FosB from Staphylococcus aureus at 1.80 Angstrom Resolution with L-Cysteine-Cys9 Disulfide | | Descriptor: | CYSTEINE, Metallothiol transferase FosB, SULFATE ION | | Authors: | Cook, P.D, Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Armstrong, R.N. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
4NB2
 
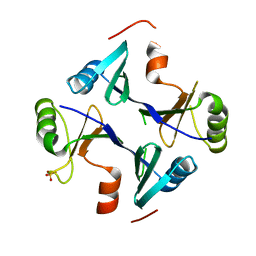 | | Crystal Structure of FosB from Staphylococcus aureus at 1.89 Angstrom Resolution - Apo structure | | Descriptor: | Metallothiol transferase FosB, SULFATE ION | | Authors: | Cook, P.D, Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Armstrong, R.N. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
4NB3
 
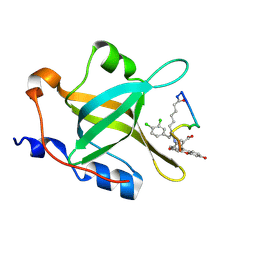 | | Crystal structure of RPA70N in complex with a 3,4 dichlorophenylalanine ATRIP derived peptide | | Descriptor: | 3,4 dichlorophenylalanine ATRIP derived peptide, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Souza-Fagundes, E.M, Luzwik, J.W, Cortez, D, Olejniczak, O.T, Waterson, A.G, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of a Potent Stapled Helix Peptide That Binds to the 70N Domain of Replication Protein A.
J.Med.Chem., 57, 2014
|
|
4NB4
 
 | | Pantothenamide-bound Pantothenate kinase from Staphylococcus aureus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, N-[2-(1,3-benzodioxol-5-yl)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Smil, D, Park, H.W. | | Deposit date: | 2013-10-22 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural characterization of a new N-substituted pantothenamide bound to pantothenate kinases from Klebsiella pneumoniae and Staphylococcus aureus.
Proteins, 82, 2014
|
|
4NB5
 
 | |
4NB6
 
 | | Crystal structure of the ligand binding domain of RORC with T0901317 | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, Nuclear receptor ROR-gamma | | Authors: | Hymowitz, S.G, Boenig-de Leon, G. | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based design of substituted hexafluoroisopropanol-arylsulfonamides as modulators of RORc.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4NB7
 
 | | Crystal Structure of Two-Domain Laccase from Streptomyces LIvidans AC1709 in complex with azide after 180 min soaking | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, AZIDE ION, ... | | Authors: | Gabdulkhakov, A, Tischenko, S, Yurevich, L, Lisov, A, Leontievsky, A. | | Deposit date: | 2013-10-23 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Two-Domain Laccase from Streptomyces Lividans AC1709 in complex with azide after 180 min soaking
To be Published
|
|
4NB8
 
 | | Oxygenase with Ile262 replaced by Leu and ferredoxin complex of carbazole 1,9a-dioxygenase | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin CarAc, ... | | Authors: | Ashikawa, Y, Usami, Y, Inoue, K, Nojiri, H. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of the divergent oxygenation reactions catalyzed by the rieske nonheme iron oxygenase carbazole 1,9a-dioxygenase.
Appl.Environ.Microbiol., 80, 2014
|
|
4NB9
 
 | | Oxygenase with Ile262 replaced by Val and ferredoxin complex of carbazole 1,9a-dioxygenase | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin CarAc, ... | | Authors: | Ashikawa, Y, Usami, Y, Inoue, K, Nojiri, H. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of the divergent oxygenation reactions catalyzed by the rieske nonheme iron oxygenase carbazole 1,9a-dioxygenase.
Appl.Environ.Microbiol., 80, 2014
|
|
4NBA
 
 | | Carbazole-bound oxygenase with Ile262 replaced by Val and ferredoxin complex of carbazole 1,9a-dioxygenase | | Descriptor: | 9H-CARBAZOLE, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ashikawa, Y, Usami, Y, Inoue, K, Nojiri, H. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the divergent oxygenation reactions catalyzed by the rieske nonheme iron oxygenase carbazole 1,9a-dioxygenase.
Appl.Environ.Microbiol., 80, 2014
|
|
4NBB
 
 | | Carbazole- and oxygen-bound oxygenase with Ile262 replaced by Val and ferredoxin complex of carbazole 1,9a-dioxygenase | | Descriptor: | 9H-CARBAZOLE, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ashikawa, Y, Usami, Y, Inoue, K, Nojiri, H. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of the divergent oxygenation reactions catalyzed by the rieske nonheme iron oxygenase carbazole 1,9a-dioxygenase.
Appl.Environ.Microbiol., 80, 2014
|
|
4NBC
 
 | | Oxygenase with Phe275 replaced by Trp and ferredoxin complex of carbazole 1,9a-dioxygenase (form1) | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin CarAc, ... | | Authors: | Ashikawa, Y, Usami, Y, Inoue, K, Nojiri, H. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Structural basis of the divergent oxygenation reactions catalyzed by the rieske nonheme iron oxygenase carbazole 1,9a-dioxygenase.
Appl.Environ.Microbiol., 80, 2014
|
|
