8BVX
 
 | |
2QOS
 
 | | Crystal structure of complement protein C8 in complex with a peptide containing the C8 binding site on C8 | | Descriptor: | Complement component 8, gamma polypeptide, Complement component C8 alpha chain | | Authors: | Lovelace, L.L, Chiswell, B, Slade, D.J, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2007-07-20 | | Release date: | 2007-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of complement protein C8gamma in complex with a peptide containing the C8gamma binding site on C8alpha: Implications for C8gamma ligand binding.
Mol.Immunol., 45, 2008
|
|
4NXH
 
 | |
3HLP
 
 | |
7DTZ
 
 | | FGFR4 complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[5-[[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]methoxy]pyrimidin-2-yl]amino]-3-methyl-phenyl]-2-fluoranyl-prop-2-enamide, SULFATE ION | | Authors: | Chen, X.J, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-01-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Investigation of Covalent Warheads in the Design of 2-Aminopyrimidine-based FGFR4 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
5B1R
 
 | | Crystal structure of mouse CD72a CTLD | | Descriptor: | ACETATE ION, B-cell differentiation antigen CD72, GLYCEROL | | Authors: | Shinagawa, K, Numoto, N, Tsubata, T, Ito, N. | | Deposit date: | 2015-12-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | CD72 negatively regulates B lymphocyte responses to the lupus-related endogenous toll-like receptor 7 ligand Sm/RNP
J.Exp.Med., 213, 2016
|
|
1GQH
 
 | | Quercetin 2,3-dioxygenase in complex with the inhibitor kojic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-HYDROXY-2-(HYDROXYMETHYL)-4H-PYRAN-4-ONE, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2001-11-23 | | Release date: | 2002-06-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional Analysis of the Copper-Dependent Quercetin 2,3-Dioxygenase.1.Ligand-Induced Coordination Changes Probed by X-Ray Crystallography: Inhibition, Ordering Effect and Mechanistic Insights
Biochemistry, 41, 2002
|
|
8C18
 
 | | Solution structure of carotenoid-binding protein AstaPo1 in complex with astaxanthin | | Descriptor: | ASTAXANTHIN, Astaxanthin binding fasciclin family protein | | Authors: | Kornilov, F.D, Savitskaya, A.G, Slonimskiy, Y.B, Goncharuk, S.A, Sluchanko, N.N, Mineev, K.S. | | Deposit date: | 2022-12-20 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the ligand promiscuity of the neofunctionalized, carotenoid-binding fasciclin domain protein AstaP.
Commun Biol, 6, 2023
|
|
7P7A
 
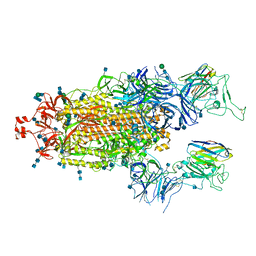 | | SARS-CoV-2 spike protein in complex with sybody#68 in a 2up/1flexible conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.76 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P77
 
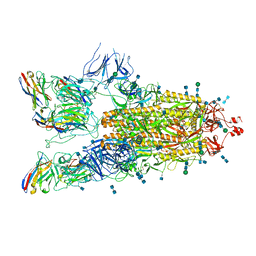 | | SARS-CoV-2 spike protein in complex with sybody#15 and sybody#68 in a 3up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P78
 
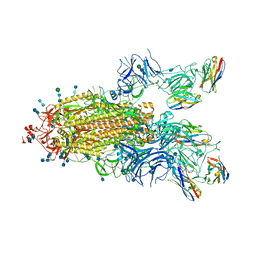 | | SARS-CoV-2 spike protein in complex with sybody#15 and sybody#68 in a 1up/1up-out/1down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P79
 
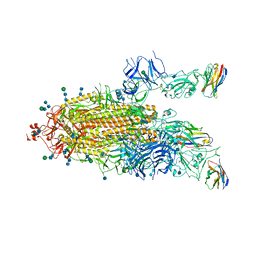 | | SARS-CoV-2 spike protein in complex with sybodyb#15 in a 1up/1up-out/1down conformation. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P7B
 
 | | SARS-CoV-2 spike protein in complex with sybody no68 in a 1up/2down conformation | | Descriptor: | Spike glycoprotein | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
3MCV
 
 | | Structure of PTR1 from Trypanosoma brucei in ternary complex with 2,4-diamino-5-[2-(2,5-dimethoxyphenyl)ethyl]thieno[2,3-d]-pyrimidine and NADP+ | | Descriptor: | 5-[2-(2,5-dimethoxyphenyl)ethyl]thieno[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2010-03-29 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structures of Trypanosoma brucei pteridine reductase ligand complexes inform on the placement of new molecular entities in the active site of a potential drug target.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1R4S
 
 | | URATE OXIDASE FROM ASPERGILLUS FLAVUS COMPLEXED WITH ITS INHIBITOR 9-METHYL URIC ACID | | Descriptor: | 9-METHYL URIC ACID, CYSTEINE, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Prange, T. | | Deposit date: | 2003-10-08 | | Release date: | 2004-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SEJ
 
 | | Crystal Structure of Dihydrofolate Reductase-Thymidylate Synthase from Cryptosporidium hominis Bound to 1843U89/NADPH/dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, ... | | Authors: | Anderson, A.C. | | Deposit date: | 2004-02-17 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Two crystal structures of dihydrofolate reductase-thymidylate synthase from Cryptosporidium hominis reveal protein-ligand interactions including a structural basis for observed antifolate resistance.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
4ZWC
 
 | | Crystal structure of maltose-bound human GLUT3 in the outward-open conformation at 2.6 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
4ZW9
 
 | | Crystal structure of human GLUT3 bound to D-glucose in the outward-occluded conformation at 1.5 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
3MUR
 
 | | Crystal Structure of the C92U mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
4CDA
 
 | | Spectroscopically-validated structure of ferric cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C', HEME C, SULFATE ION | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-10-30 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3MUM
 
 | | Crystal Structure of the G20A mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), G20A mutant c-di-GMP Riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
6UHO
 
 | | Crystal Structure of C148 mGFP-cDNA-2 | | Descriptor: | C148 mGFP-cDNA-2, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
3MUV
 
 | | Crystal Structure of the G20A/C92U mutant c-di-GMP riboswith bound to c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, G20A/C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
5LSP
 
 | | 107_A07 Fab in complex with fragment of the Met receptor | | Descriptor: | 107_A07 Fab heavy chain, 107_A07 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DiCara, D, Chirgadze, D.Y, Pope, A, Karatt-Vellatt, A, Winter, A, van den Heuvel, J, Gherardi, E, McCafferty, J. | | Deposit date: | 2016-09-05 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Characterization and structural determination of a new anti-MET function-blocking antibody with binding epitope distinct from the ligand binding domain.
Sci Rep, 7, 2017
|
|
2AXT
 
 | | Crystal Structure of Photosystem II from Thermosynechococcus elongatus | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Loll, B, Kern, J, Saenger, W, Zouni, A, Biesiadka, J. | | Deposit date: | 2005-09-06 | | Release date: | 2005-12-27 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Towards complete cofactor arrangement in the 3.0 A resolution structure of photosystem II
NATURE, 438, 2005
|
|
