6CTR
 
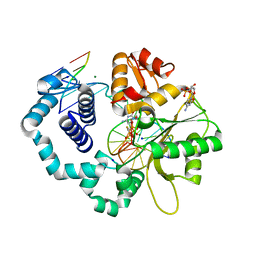 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CHF (R & S isomers), beta, gamma dCTP analogue | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-deoxy-5'-O-[(R)-{[(R)-[(R)-fluoro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, 2'-deoxy-5'-O-[(R)-{[(R)-[(S)-fluoro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CTV
 
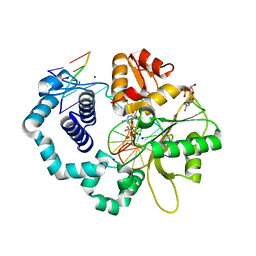 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CF2, beta, gamma dCTP analogue | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-deoxy-5'-O-[(R)-{[(R)-[difluoro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, CHLORIDE ION, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CTX
 
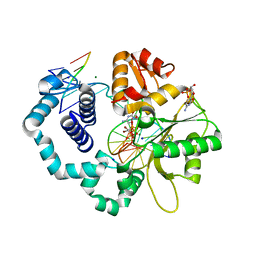 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CBr2, beta, gamma dCTP analogue | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-deoxy-5'-O-[(R)-{[(R)-[dibromo(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, CHLORIDE ION, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6BEM
 
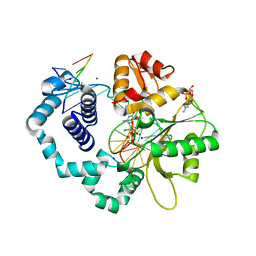 | | Ternary complex crystal structure of DNA polymerase Beta with S-isomer of beta-gamma-CHCL-dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 5'-O-[(R)-{[(R)-[(S)-chloro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-2'-deoxycytidine, CHLORIDE ION, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2017-10-25 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6BEL
 
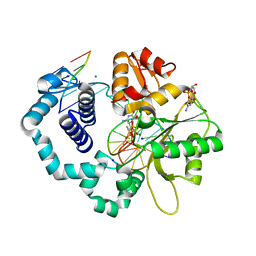 | | Ternary complex crystal structure of DNA polymerase Beta with R-isomer of beta-gamma-CHF-dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-deoxy-5'-O-[(R)-{[(R)-[(R)-fluoro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, CHLORIDE ION, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2017-10-25 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CR7
 
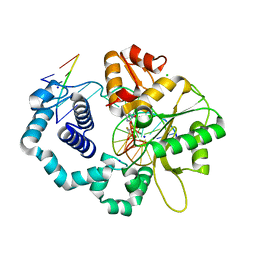 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CHF, beta, gamma dATP analogue | | Descriptor: | 9-{2-deoxy-5-O-[(R)-{[(R)-[(R)-fluoro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-alpha-D-erythro-pentofuranosyl}-9H-purin-6-amine, CHLORIDE ION, DNA polymerase beta, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CRB
 
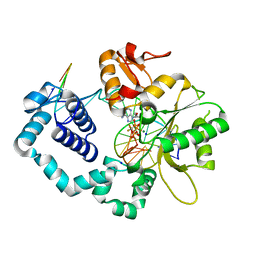 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CF2, beta, gamma dATP analogue | | Descriptor: | 9-{2-deoxy-5-O-[(S)-{[(S)-[difluoro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-alpha-D-erythro-pentofuranosyl}-9H-purin-6-amine, DNA polymerase beta, Downstream Primer Strand, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CR4
 
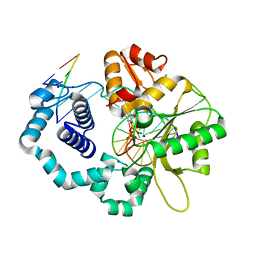 | |
8UDK
 
 | | Human Mitochondrial DNA Polymerase gamma R853A Ternary Complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (24-MER), DNA (28-MER), ... | | Authors: | Park, J, Herrmann, G.K, Yin, Y.W. | | Deposit date: | 2023-09-28 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | An interaction network in the polymerase active site is a prerequisite for Watson-Crick base pairing in Pol gamma.
Sci Adv, 10, 2024
|
|
6CR5
 
 | |
6CR6
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CH-CH3, beta, gamma dATP analogue | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy({(R)-hydroxy[(1S)-1-phosphonoethyl]phosphoryl}oxy)phosphoryl]adenosine, DNA polymerase beta, Downstream Primer Strand, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CRC
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CCL2, beta, gamma dATP analogue | | Descriptor: | 2'-deoxy-5'-O-[(R)-{[(R)-[dichloro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]adenosine, DNA polymerase beta, Downstream Primer Strand, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CR9
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CFCL, beta, gamma dATP analogue | | Descriptor: | 9-{5-O-[(R)-{[(R)-[(S)-chloro(fluoro)phosphonomethyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-2-deoxy-alpha-D-threo-pentofuranosyl}-9H-purin-6-amine, CHLORIDE ION, DNA polymerase beta, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CR8
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CHCL (R & S isomers), beta, gamma dATP analogue | | Descriptor: | 5'-O-[(R)-{[(R)-[(R)-chloro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-2'-deoxyadenosine, CHLORIDE ION, DNA polymerase beta, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
4F3E
 
 | |
2G3R
 
 | | Crystal Structure of 53BP1 tandem tudor domains at 1.2 A resolution | | Descriptor: | SULFATE ION, Tumor suppressor p53-binding protein 1 | | Authors: | Lee, J, Botuyan, M.V, Thompson, J.R, Mer, G. | | Deposit date: | 2006-02-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis for the Methylation State-Specific Recognition of Histone H4-K20 by 53BP1 and Crb2 in DNA Repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
7CUX
 
 | | Crystal structure of human Schlafen 5 N'-terminal domain (SLFN5-N) involved in ssRNA cleaving and DNA binding | | Descriptor: | Schlafen family member 5, ZINC ION | | Authors: | Yang, J.Y, Luo, M, Ou, J.Y, Wang, Z.W, Gao, S. | | Deposit date: | 2020-08-25 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.29477072 Å) | | Cite: | Crystal structure of human Schlafen 5 N'-terminal domain (SLFN5-N) involved in ssRNA cleaving and DNA binding
To Be Published
|
|
5NBL
 
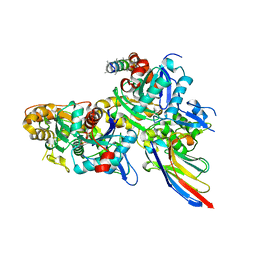 | | Crystal structure of the Arp4-N-actin(APO-state) heterodimer bound by a nanobody | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Knoll, K.R, Eustermann, S, Hopfner, K.P. | | Deposit date: | 2017-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3T72
 
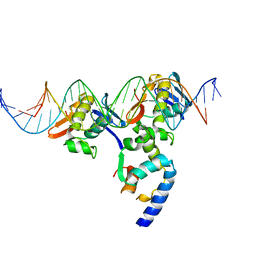 | | PhoB(E)-Sigma70(4)-(RNAP-Betha-flap-tip-helix)-DNA Transcription Activation Sub-Complex | | Descriptor: | PHO BOX DNA (STRAND 1), PHO BOX DNA (STRAND 2), Phosphate regulon transcriptional regulatory protein phoB, ... | | Authors: | Blanco, A.G, Canals, A, Bernues, J, Sola, M, Coll, M. | | Deposit date: | 2011-07-29 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.33 Å) | | Cite: | The structure of a transcription activation subcomplex reveals how sigma (70) is recruited to PhoB promoters.
Embo J., 30, 2011
|
|
4WLS
 
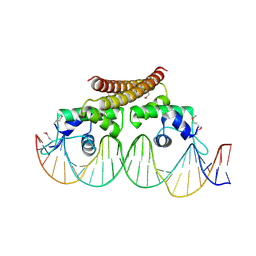 | | Crystal structure of the metal-free (repressor) form of E. Coli CUER, a copper efflux regulator, bound to COPA promoter DNA | | Descriptor: | COPA PROMOTER DNA NON-TEMPLATE STRAND, COPA PROMOTER DNA NON-TEMPLATE STRAND (ALTERNATE CONFORMATION), COPA PROMOTER DNA TEMPLATE STRAND, ... | | Authors: | Philips, S.J, Canalizo-Hernandez, M, Mondragon, A, O'Halloran, T.V. | | Deposit date: | 2014-10-08 | | Release date: | 2015-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Allosteric transcriptional regulation via changes in the overall topology of the core promoter.
Science, 349, 2015
|
|
5NBM
 
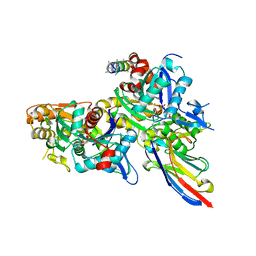 | | Crystal structure of the Arp4-N-actin(ATP-state) heterodimer bound by a nanobody | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Knoll, K.R, Eustermann, S, Hopfner, K.P. | | Deposit date: | 2017-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6WX5
 
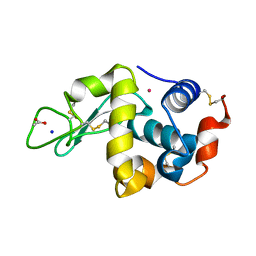 | | Adducts formed after 3 weeks in the reaction of chlorido[chlorido(2,2'-((2-([2,2':6',2''-Terpyridin]-4'-yloxy)ethyl)azanediyl)bis(ethan-1-ol))platinum(II)] with HEWL | | Descriptor: | ACETATE ION, Lysozyme, PLATINUM (II) ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2020-05-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mustards-Derived Terpyridine-Platinum Complexes as Anticancer Agents: DNA Alkylation vs Coordination.
Inorg.Chem., 60, 2021
|
|
5EDW
 
 | |
4FE4
 
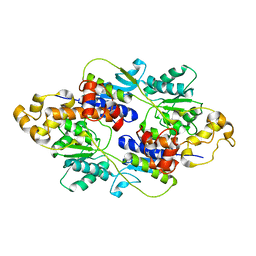 | | Crystal structure of apo E. coli XylR | | Descriptor: | Xylose operon regulatory protein | | Authors: | Schumacher, M.A, Ni, L. | | Deposit date: | 2012-05-29 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structures of the Escherichia coli transcription activator and regulator of diauxie, XylR: an AraC DNA-binding family member with a LacI/GalR ligand-binding domain.
Nucleic Acids Res., 41, 2013
|
|
6MIL
 
 | | Crystal structure of AF9 YEATS domain in complex with histone H3K9bu | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histone H3K9bu, MALONATE ION, ... | | Authors: | Vann, K.R, Klein, B.J, Kutateladze, T.G. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
