1KVF
 
 | |
1GX5
 
 | |
1GX6
 
 | |
6F0S
 
 | |
6F0T
 
 | | Crystal structure of Pizza6-SFW | | Descriptor: | GLYCEROL, Pizza6-SFW | | Authors: | Noguchi, H, De Zitter, E, Van Meervelt, L, Voet, A.R.D. | | Deposit date: | 2017-11-20 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Design of tryptophan-containing mutants of the symmetrical Pizza protein for biophysical studies.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
6QAM
 
 | |
8QM0
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AmiA in complex with Peptide 5 | | Descriptor: | ALA-LYS-THR-ILE-LYS-ILE-THR-GLN-THR-ARG, Oligopeptide-binding protein AmiA | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
1C3T
 
 | |
8U89
 
 | | The structure of the PP2A-B56Delta holoenzyme mutant - E197K | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Wu, C.G, Xing, Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | B56 delta long-disordered arms form a dynamic PP2A regulation interface coupled with global allostery and Jordan's syndrome mutations.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1D7M
 
 | | COILED-COIL DIMERIZATION DOMAIN FROM CORTEXILLIN I | | Descriptor: | CORTEXILLIN I | | Authors: | Burkhard, P, Kammerer, R.A, Steinmetz, M.O, Bourenkov, G.P, Aebi, U. | | Deposit date: | 1999-10-19 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The coiled-coil trigger site of the rod domain of cortexillin I unveils a distinct network of interhelical and intrahelical salt bridges.
Structure Fold.Des., 8, 2000
|
|
8U1X
 
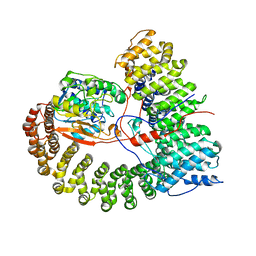 | | The structure of the PP2A-B56Delta holoenzyme mutant - E197K | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Wu, C.G, Xing, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | B56 delta long-disordered arms form a dynamic PP2A regulation interface coupled with global allostery and Jordan's syndrome mutations.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2P11
 
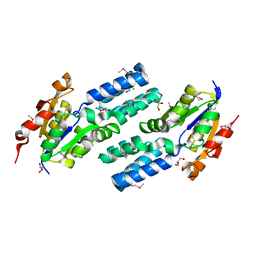 | |
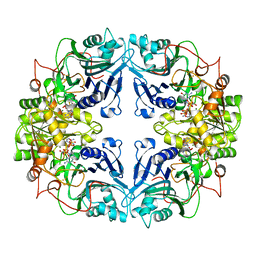
1AO0
 
 | |
2QUZ
 
 | |
5IPF
 
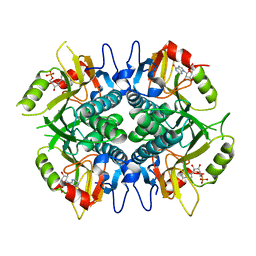 | | Crystal structure of Hypoxanthine-guanine phosphoribosyltransferase from Schistosoma mansoni in complex with IMP | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase (HGPRT), INOSINIC ACID | | Authors: | Romanello, L, Torini, J.R.S, Bird, L.E, Nettleship, J.E, Owens, R.J, DeMarco, R, Pereira, H.M, Brandao-Neto, J. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In vitro and in vivo characterization of the multiple isoforms of Schistosoma mansoni hypoxanthine-guanine phosphoribosyltransferases.
Mol. Biochem. Parasitol., 229, 2019
|
|
9BQ0
 
 | |
6NBR
 
 | |
3D2O
 
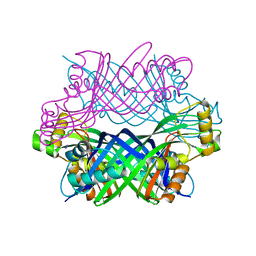 | | Crystal Structure of Manganese-metallated GTP Cyclohydrolase Type IB | | Descriptor: | AZIDE ION, CHLORIDE ION, LITHIUM ION, ... | | Authors: | Swairjo, M.A. | | Deposit date: | 2008-05-08 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Zinc-independent folate biosynthesis: genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB
J.Bacteriol., 191, 2009
|
|
2JO4
 
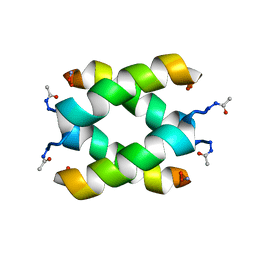 | | Tetrameric structure of KIA7 peptide | | Descriptor: | KIA7 | | Authors: | Lopez de la Osa, J, Gonzalez, C, Laurents, D.V, Chakrabartty, A, Bateman, D.A. | | Deposit date: | 2007-02-21 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Getting specificity from simplicity in putative proteins from the prebiotic earth.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8DL3
 
 | | Crystal structure of the human queuine salvage enzyme DUF2419, complexed with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, Queuosine salvage protein | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-07-06 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
2JO5
 
 | | Tetrameric structure of KIA7F peptide | | Descriptor: | KIA7F | | Authors: | Lopez de la Osa, J, Gonzalez, C, Laurents, D.V, Chakrabartty, A, Bateman, D.A. | | Deposit date: | 2007-02-21 | | Release date: | 2007-09-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Getting specificity from simplicity in putative proteins from the prebiotic earth.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6P78
 
 | | queuine lyase from Clostridium spiroforme bound to SAM and queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5S)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-1,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, IRON/SULFUR CLUSTER, Queuine lyase, ... | | Authors: | Almo, S.C, Grove, T.L. | | Deposit date: | 2019-06-05 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.726 Å) | | Cite: | Discovery of novel bacterial queuine salvage enzymes and pathways in human pathogens.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OP5
 
 | |
8A42
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AmiA in complex with an unknown peptide | | Descriptor: | Oligopeptide-binding protein AmiA, Unknown peptide | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2022-06-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
7K12
 
 | | ACMSD in complex with diflunisal | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, CITRIC ACID, ... | | Authors: | Yang, Y, Liu, A. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Diflunisal Derivatives as Modulators of ACMS Decarboxylase Targeting the Tryptophan-Kynurenine Pathway.
J.Med.Chem., 64, 2021
|
|
