6WJD
 
 | |
6XIQ
 
 | | Cryo-EM Structure of K63R Ubiquitin Mutant Ribosome under Oxidative Stress | | Descriptor: | 18S ribosomal RNA, 35S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhou, Y, Bartesaghi, A, Silva, G.M. | | Deposit date: | 2020-06-21 | | Release date: | 2020-08-26 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural impact of K63 ubiquitin on yeast translocating ribosomes under oxidative stress.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W2T
 
 | | Structure of the Cricket Paralysis Virus 5-UTR IRES (CrPV 5-UTR-IRES) bound to the small ribosomal subunit in the closed state (Class 2) | | Descriptor: | 18S rRNA, CrPV 5'-UTR IRES, Eukaryotic translation initiation factor 3 subunit A, ... | | Authors: | Neupane, R, Pisareva, V, Rodriguez, C.F, Pisarev, A, Fernandez, I.S. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A complex IRES at the 5'-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate.
Elife, 9, 2020
|
|
6Y0G
 
 | | Structure of human ribosome in classical-PRE state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
6Y53
 
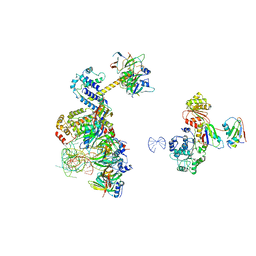 | | human 17S U2 snRNP low resolution part | | Descriptor: | HIV Tat-specific factor 1, Probable ATP-dependent RNA helicase DDX46, Small nuclear ribonucleoprotein E, ... | | Authors: | Zhang, Z, Will, C.L, Bertram, K, Luehrmann, R, Stark, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Molecular architecture of the human 17S U2 snRNP.
Nature, 583, 2020
|
|
6YAM
 
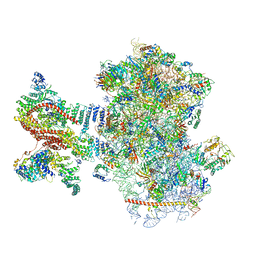 | | Mammalian 48S late-stage translation initiation complex (LS48S+eIF3 IC) with beta-globin mRNA | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein eS1, 40S ribosomal protein eS10, ... | | Authors: | Bochler, A, Simonetti, A, Guca, E, Hashem, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-04-08 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Insights into the Mammalian Late-Stage Initiation Complexes.
Cell Rep, 31, 2020
|
|
6Y57
 
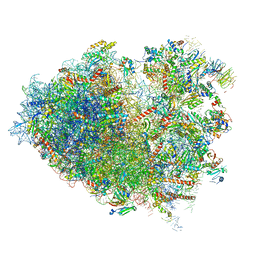 | | Structure of human ribosome in hybrid-PRE state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-25 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
5T2A
 
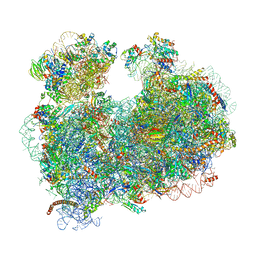 | | CryoEM structure of the Leishmania donovani 80S ribosome at 2.9 Angstrom resolution | | Descriptor: | 18S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Zhang, X, Lai, M, Zhou, Z.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-01-25 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
5T62
 
 | | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3-Tif6-Lsg1 Complex | | Descriptor: | 25S Ribosomal RNA, 5.8S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Malyutin, A.G, Musalgaonkar, S, Patchett, S, Frank, J, Johnson, A.W. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
EMBO J., 36, 2017
|
|
5T2C
 
 | | CryoEM structure of the human ribosome at 3.6 Angstrom resolution | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Zhang, X, Lai, M, Zhou, Z.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-01-25 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
5T6R
 
 | | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3 Complex | | Descriptor: | 25S Ribosomal RNA, 5.8S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Malyutin, A.G, Musalgaonkar, S, Patchett, S, Frank, J, Johnson, A.W. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
EMBO J., 36, 2017
|
|
5TGM
 
 | | Crystal structure of the S.cerevisiae 80S ribosome in complex with the A-site bound aminoacyl-tRNA analog ACCA-Pro | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3'-amino-3'-deoxyadenosine 5'-(dihydrogen phosphate), ... | | Authors: | Melnikov, S, Mailliot, J, Yusupov, M. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-18 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular insights into protein synthesis with proline residues.
EMBO Rep., 17, 2016
|
|
2EKE
 
 | |
5UDH
 
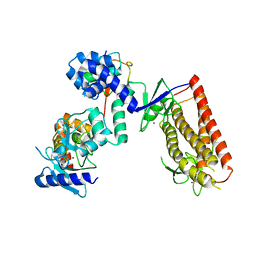 | | HHARI/ARIH1-UBCH7~Ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, Ubiquitin C variant, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Miller, D.J, Schulman, B.A. | | Deposit date: | 2016-12-27 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural Studies of HHARI/UbcH7Ub Reveal Unique E2Ub Conformational Restriction by RBR RING1.
Structure, 25, 2017
|
|
2C9W
 
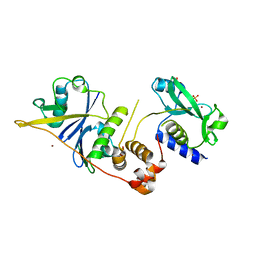 | | CRYSTAL STRUCTURE OF SOCS-2 IN COMPLEX WITH ELONGIN-B AND ELONGIN-C AT 1.9A RESOLUTION | | Descriptor: | NICKEL (II) ION, SULFATE ION, SUPPRESSOR OF CYTOKINE SIGNALING 2, ... | | Authors: | Debreczeni, J.E, Bullock, A, Amos, A, Savitsky, P, Barr, A, Burgess, N, Sundstrom, M, Weigelt, J, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 103, 2006
|
|
7F5S
 
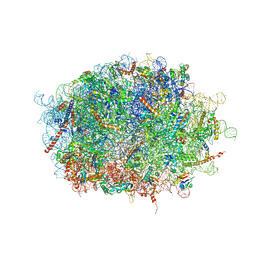 | | human delta-METTL18 60S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Takahashi, M, Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | METTL18-mediated histidine methylation of RPL3 modulates translation elongation for proteostasis maintenance.
Elife, 11, 2022
|
|
7EVO
 
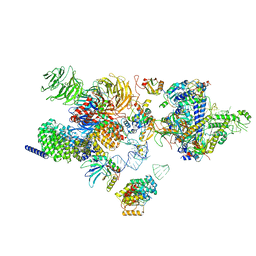 | | The cryo-EM structure of the human 17S U2 snRNP | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, RNA helicase, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome.
Nat.Struct.Mol.Biol., 2024
|
|
4P5O
 
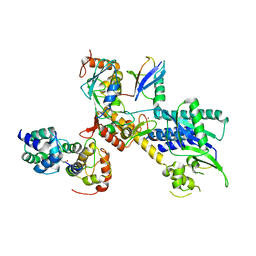 | |
7JQC
 
 | | SARS-CoV-2 Nsp1, CrPV IRES and rabbit 40S ribosome complex | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S24, 40S ribosomal protein S26, ... | | Authors: | Yuan, S, Xiong, Y. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA.
Mol.Cell, 80, 2020
|
|
7JQB
 
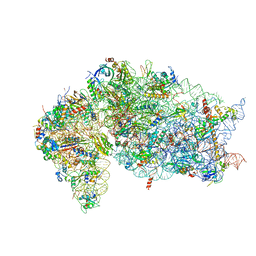 | | SARS-CoV-2 Nsp1 and rabbit 40S ribosome complex | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S24, 40S ribosomal protein S26, ... | | Authors: | Yuan, S, Xiong, Y. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA.
Mol.Cell, 80, 2020
|
|
4Q5H
 
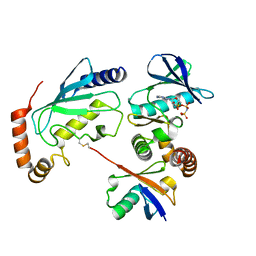 | |
7K5I
 
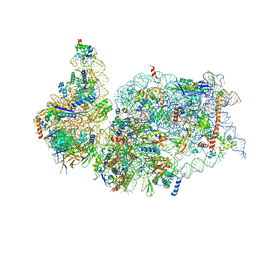 | | SARS-COV-2 nsp1 in complex with human 40S ribosome | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Wang, L, Shi, M, Wu, H. | | Deposit date: | 2020-09-16 | | Release date: | 2020-10-14 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism.
Biorxiv, 2020
|
|
4Q5E
 
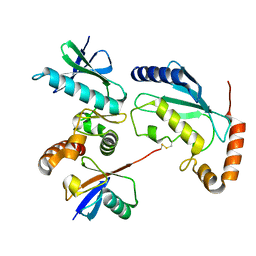 | |
7K5J
 
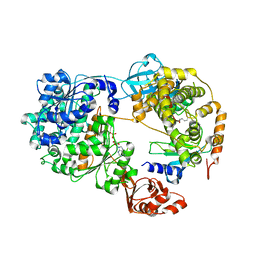 | | Structure of an E1-E2-ubiquitin thioester mimetic | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Yuan, L, Lv, Z, Olsen, S.K. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Crystal structures of an E1-E2-ubiquitin thioester mimetic reveal molecular mechanisms of transthioesterification.
Nat Commun, 12, 2021
|
|
7K6Q
 
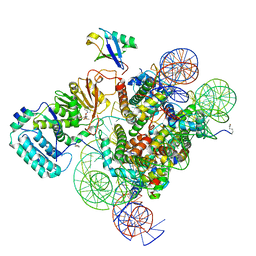 | | Active state Dot1 bound to the H4K16ac nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Valencia-Sanchez, M.I, De Ioannes, P.E, Miao, W, Truong, D.M, Lee, R, Armache, J.-P, Boeke, J.D, Armache, K.-J. | | Deposit date: | 2020-09-21 | | Release date: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation.
Science, 371, 2021
|
|
