4U95
 
 | | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, DARPin, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pos, K.M. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB
eLife, 3, 2014
|
|
4UAN
 
 | |
4TX6
 
 | | AfChiA1 in complex with compound 1 | | Descriptor: | 3-(2-methoxyphenyl)-6-methyl[1,2]oxazolo[5,4-d]pyrimidin-4(5H)-one, Class III chitinase ChiA1, PHOSPHATE ION | | Authors: | van Aalten, D.M.F. | | Deposit date: | 2014-07-02 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Screening-based discovery of Aspergillus fumigatus plant-type chitinase inhibitors
FEBS Lett, 588, 2014
|
|
4TXN
 
 | | Crystal structure of uridine phosphorylase from Schistosoma mansoni in complex with 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, SULFATE ION, Uridine phosphorylase | | Authors: | Marinho, A, Torini, J, Romanello, L, Cassago, A, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2014-07-03 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of two Schistosoma mansoni uridine phosphorylases isoforms suggests the emergence of a protein with a non-canonical function.
Biochimie, 125, 2016
|
|
5POH
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N11029a | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, N-[2-(phenylamino)ethyl]acetamide, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4TY8
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 6-methyl-2H-chromen-2-one, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4UB7
 
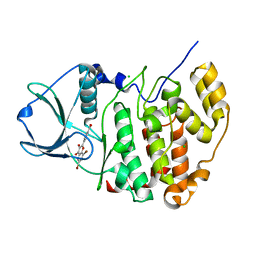 | | High-salt structure of protein kinase CK2 catalytic subunit with 4'-carboxy-6,8-bromo-flavonol (FLC26) showing an extreme distortion of the ATP-binding loop combined with a pi-halogen bond | | Descriptor: | 4-(6,8-dibromo-3-hydroxy-4-oxo-4H-chromen-2-yl)benzoic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Niefind, K, Bischoff, N, Guerra, B, Golub, A, Issinger, O.-G. | | Deposit date: | 2014-08-12 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Note of Caution on the Role of Halogen Bonds for Protein Kinase/Inhibitor Recognition Suggested by High- And Low-Salt CK2 alpha Complex Structures.
Acs Chem.Biol., 10, 2015
|
|
4UBK
 
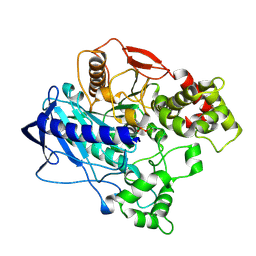 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 7.40 MGy at 100K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
4UBV
 
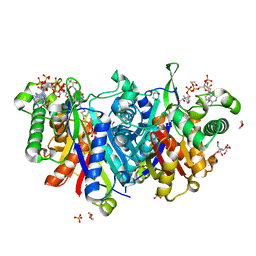 | |
4TYT
 
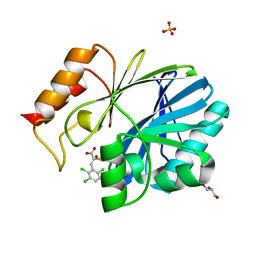 | | Crystal Structure of BcII metallo-beta-lactamase in complex with ML302F | | Descriptor: | (2Z)-2-sulfanyl-3-(2,3,6-trichlorophenyl)prop-2-enoic acid, Beta-lactamase 2, GLYCEROL, ... | | Authors: | Brem, J, van Berkel, S.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2014-07-09 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Rhodanine hydrolysis leads to potent thioenolate mediated metallo-beta-lactamase inhibition.
Nat.Chem., 6, 2014
|
|
4TZ1
 
 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose | | Descriptor: | Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4U1Y
 
 | | Full length GluA2-FW-(R,R)-2b complex | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.8999 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U2A
 
 | | Structure of a lectin from the seeds of Vatairea macrocarpa complexed with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, CITRIC ACID, ... | | Authors: | Sousa, B.L, Silva-Filho, J.C, Kumar, P, Lyskowski, A, Bezerra, G.A, Delatorre, P, Rocha, B.A.M, Nagano, C.S, Gruber, K, Cavada, B.S. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | High-resolution structure of a new Tn antigen-binding lectin from Vatairea macrocarpa and a comparative analysis of Tn-binding legume lectins.
Int.J.Biochem.Cell Biol., 59C, 2014
|
|
4U2C
 
 | | Crystal structure of dienelactone hydrolase A-6 variant (S7T, A24V, Q35H, F38L, Q110L, C123S, Y145C, E199G and S208G) at 1.95 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4U2D
 
 | | Crystal structure of dienelactone hydrolase S-2 variant (Q35H, F38L, Q110L, C123S, Y137C, Y145C, N154D, E199G, S208G and G211D) at 1.67 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4U2G
 
 | | Crystal structure of dienelactone hydrolase B-4 variant (Q35H, F38L, Y64H, Q76L, Q110L, C123S, Y137C, A141V, Y145C, N154D, E199G, S208G, G211D, S233G and 237Q) at 1.80 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4UDJ
 
 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.60 Angstrom in complex with beta-D-mannopyranose and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UFI
 
 | | Mouse Galactocerebrosidase complexed with aza-galacto-fagomine AGF | | Descriptor: | (3R,4S,5R)-3-(hydroxymethyl)-1,2-diazinane-4,5-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hill, C.H, Viuff, A.H, Spratley, S.J, Salamone, S, Christensen, S.H, Read, R.J, Moriarty, N.W, Jensen, H.H, Deane, J.E. | | Deposit date: | 2015-03-17 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Azasugar Inhibitors as Pharmacological Chaperones for Krabbe Disease.
Chem.Sci., 6, 2015
|
|
4TUZ
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with alpha-zearalenol | | Descriptor: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, 1,2-ETHANEDIOL, Estrogen receptor, ... | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2014-06-25 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural perspective on nuclear receptors as targets of environmental compounds.
Acta Pharmacol. Sin., 36, 2015
|
|
4TV8
 
 | | Tubulin-Maytansine complex | | Descriptor: | (3beta,4beta,5beta,10beta,11E,13E)-maytansine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Bargsten, K, Diaz, J.F, Marsh, M, Cuevas, C, Liniger, M, Neuhaus, C, Andreu, J.M, Altmann, K.H, Steinmetz, M.O. | | Deposit date: | 2014-06-26 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TWA
 
 | |
4U2H
 
 | |
8REN
 
 | | Crystal structure of reduced ThyX | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent thymidylate synthase, ... | | Authors: | Pecqueur, L, Hamdane, D. | | Deposit date: | 2023-12-12 | | Release date: | 2025-01-01 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.139 Å) | | Cite: | Structural Plasticity of Flavin-Dependent Thymidylate Synthase Controlled by the Enzyme Redox State.
Biomolecules, 15, 2025
|
|
8REP
 
 | | Crystal structure of oxidized ThyX-Y91F mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent thymidylate synthase, ... | | Authors: | Pecqueur, L, Hamdane, D. | | Deposit date: | 2023-12-12 | | Release date: | 2025-01-01 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural Plasticity of Flavin-Dependent Thymidylate Synthase Controlled by the Enzyme Redox State.
Biomolecules, 15, 2025
|
|
8REO
 
 | | Crystal structure of reduced ThyX in complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Pecqueur, L, Hamdane, D. | | Deposit date: | 2023-12-12 | | Release date: | 2025-01-01 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Plasticity of Flavin-Dependent Thymidylate Synthase Controlled by the Enzyme Redox State.
Biomolecules, 15, 2025
|
|
