6P8Z
 
 | | Crystal structure of human KRAS G12C covalently bound to an acryloylazetidine acetamide inhibitor | | Descriptor: | 2-[5-chloro-2-cyclopropyl-3-(5-methoxy-3,4-dihydroisoquinoline-2(1H)-carbonyl)-7-methyl-1H-indol-1-yl]-N-(1-propanoylazetidin-3-yl)acetamide, CALCIUM ION, GTPase KRas, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-06-08 | | Release date: | 2019-08-28 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery ofN-(1-Acryloylazetidin-3-yl)-2-(1H-indol-1-yl)acetamides as Covalent Inhibitors of KRASG12C.
Acs Med.Chem.Lett., 10, 2019
|
|
6HH2
 
 | | Rab29 small GTPase bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-7L1 | | Authors: | Khan, A.R, McGrath, E, Waschbusch, D. | | Deposit date: | 2018-08-24 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | LRRK2 binds to the Rab32 subfamily in a GTP-dependent mannerviaits armadillo domain.
Small GTPases, 2019
|
|
6CU6
 
 | |
6HXU
 
 | | Crystal structure of Human RHOB Q63L in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rho-related GTP-binding protein RhoB | | Authors: | Soulie, S, Gence, R, Cabantous, S, Lajoie-Mazenc, I, Favre, G, Pedelacq, J.D. | | Deposit date: | 2018-10-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | A Targeted Protein Degradation Cell-Based Screening for Nanobodies Selective toward the Cellular RHOB GTP-Bound Conformation.
Cell Chem Biol, 26, 2019
|
|
6SGE
 
 | | Crystal structure of Human RHOB-GTP in complex with nanobody B6 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Nanobody B6, ... | | Authors: | Soulie, S, Gence, R, Cabantous, S, Lajoie-Mazenc, I, Favre, G, Pedelacq, J.D. | | Deposit date: | 2019-08-04 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Targeted Protein Degradation Cell-Based Screening for Nanobodies Selective toward the Cellular RHOB GTP-Bound Conformation.
Cell Chem Biol, 26, 2019
|
|
6D71
 
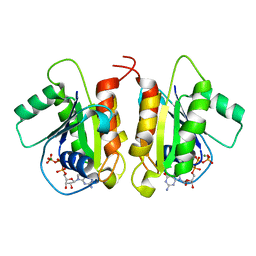 | | Crystal Structure of the Human Miro1 N-terminal GTPase bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial Rho GTPase 1 | | Authors: | Smith, K.P, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2018-04-23 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7180779 Å) | | Cite: | Insight into human Miro1/2 domain organization based on the structure of its N-terminal GTPase.
J.Struct.Biol., 212, 2020
|
|
6OB3
 
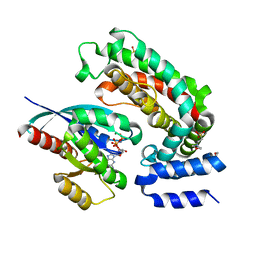 | | Crystal structure of G13D-KRAS (GMPPNP-bound) in complex with GAP-related domain (GRD) of neurofibromin (NF1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GTPase KRas, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2019-03-19 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KRAS G13D sensitivity to neurofibromin-mediated GTP hydrolysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OB2
 
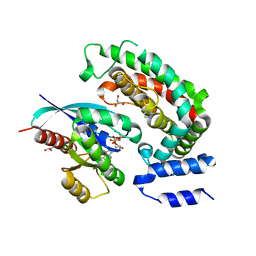 | | Crystal structure of wild-type KRAS (GMPPNP-bound) in complex with GAP-related domain (GRD) of neurofibromin (NF1) | | Descriptor: | CHLORIDE ION, GLYCEROL, GTPase KRas, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2019-03-19 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | KRAS G13D sensitivity to neurofibromin-mediated GTP hydrolysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OIM
 
 | |
6KYH
 
 | | Crystal structure of Shank3 NTD-ANK A42K mutant in complex with HRas | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Cai, Q, Zhang, M. | | Deposit date: | 2019-09-18 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Shank3 Binds to and Stabilizes the Active Form of Rap1 and HRas GTPases via Its NTD-ANK Tandem with Distinct Mechanisms.
Structure, 28, 2020
|
|
6KYK
 
 | | Crystal structure of Shank3 NTD-ANK mutant in complex with Rap1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rap-1b, ... | | Authors: | Cai, Q, Zhang, M. | | Deposit date: | 2019-09-19 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Shank3 Binds to and Stabilizes the Active Form of Rap1 and HRas GTPases via Its NTD-ANK Tandem with Distinct Mechanisms.
Structure, 28, 2020
|
|
6NAZ
 
 | | Crystal structure of DIRAS 2/3 chimera in complex with GDP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GTP-binding protein Di-Ras2,GTP-binding protein Di-Ras3,GTP-binding protein Di-Ras2, ... | | Authors: | Gilbert, Y.H, Reger, A, Sharma, R, Bast, J, Kim, C. | | Deposit date: | 2018-12-06 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.081 Å) | | Cite: | Crystal structure of DIRAS 2/3 chimera in complex with GDP
To Be Published
|
|
6IY1
 
 | | Structure of human Ras-related protein Rab11 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-11A | | Authors: | Ma, P, Li, S, Li, J. | | Deposit date: | 2018-12-12 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of human Ras-related protein Rab11
To Be Published
|
|
5QQN
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-010-382-606 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-cyclopropyl-2-(4-{[(5-methyl-1,2-oxazol-3-yl)carbamoyl]amino}-1H-pyrazol-1-yl)acetamide, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQK
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-359-835 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-{1-[2-(diethylamino)ethyl]-1H-indol-5-yl}-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQE
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-531-494 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-(5-methyl-1,2-oxazol-3-yl)-N'-[(3S)-4,4,4-trifluoro-3-hydroxy-3-(5-methylfuran-2-yl)butyl]urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQG
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-541-216 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-(5-methyl-1,2-oxazol-3-yl)-N'-[3-(4-phenylpiperazin-1-yl)propyl]urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQM
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-565-325 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-(1-benzyl-1H-pyrazol-4-yl)-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQF
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-004-412-710 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-ethyl-N'-1,3-thiazol-2-ylurea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-587-558 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-{1-[(imidazo[1,2-a]pyridin-2-yl)methyl]-1H-pyrazol-4-yl}-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQH
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-020-096-465 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-methyl-5-({[(5-methyl-1,2-oxazol-3-yl)carbamoyl]amino}methyl)thiophene-2-sulfonamide, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQI
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-178-994 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-[3-(methoxymethyl)phenyl]-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQL
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-565-301 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-(5-methyl-1,2-oxazol-3-yl)-N'-[2-(phenylsulfonyl)ethyl]urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQD
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with Z56880342 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-ethyl-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6V5L
 
 | | The HADDOCK structure model of GDP KRas in complex with its allosteric inhibitor E22 | | Descriptor: | (2R)-2-[2-(1H-indole-3-carbonyl)hydrazinyl]-2-phenylacetamide, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, X, Gupta, A.K, Prakash, P, Putkey, J.P, Gorfe, A.A. | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi target ensemble based virtual screening yields novel allosteric KRAS inhibitors at high success rate
Chemical Biology & Drug Design, 94, 2019
|
|
