8CEW
 
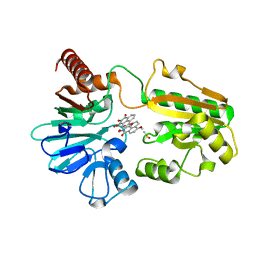 | | Crystal structure of human DNA cross-link repair 1A in complex with N-hydroxyimide inhibitor H1 | | Descriptor: | 6-methoxy-2-oxidanyl-benzo[de]isoquinoline-1,3-dione, DIMETHYL SULFOXIDE, DNA cross-link repair 1A protein, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of human DNA cross-link repair 1A in complex with N-hydroxyimide inhibitor H1
To Be Published
|
|
8CF0
 
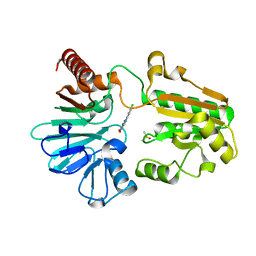 | | Crystal structure of human DNA cross-link repair 1A in complex with quinoxalinedione inhibitor H2 | | Descriptor: | 9-chloranyl-1,4-dihydropyrazino[2,3-c]quinoline-2,3-dione, DIMETHYL SULFOXIDE, DNA cross-link repair 1A protein, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of human DNA cross-link repair 1A in complex with quinoxalinedione inhibitor H2
To Be Published
|
|
1YJM
 
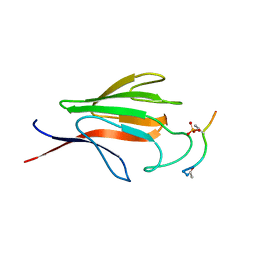 | | Crystal structure of the FHA domain of mouse polynucleotide kinase in complex with an XRCC4-derived phosphopeptide. | | Descriptor: | 12-mer peptide from DNA-repair protein XRCC4, Polynucleotide 5'-hydroxyl-kinase | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-14 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
1XEW
 
 | |
1XEX
 
 | |
1NYD
 
 | | Solution structure of DNA quadruplex GCGGTGGAT | | Descriptor: | 5'-D(*GP*CP*GP*GP*TP*GP*GP*AP*T)-3' | | Authors: | Webba da Silva, M. | | Deposit date: | 2003-02-12 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Association of DNA quadruplexes through G:C:G:C tetrads. Solution structure of d(GCGGTGGAT).
Biochemistry, 42, 2003
|
|
1V3O
 
 | | Crystal structure of d(GCGAGAGC): the DNA quadruplex structure split from the octaplex | | Descriptor: | 5'-D(*GP*(C38)P*GP*AP*GP*AP*GP*C)-3', POTASSIUM ION | | Authors: | Kondo, J, Umeda, S, Sunami, T, Takenaka, A. | | Deposit date: | 2003-11-03 | | Release date: | 2004-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats
Nucleic Acids Res., 32, 2004
|
|
1V3N
 
 | | Crystal structure of d(GCGAGAGC): the DNA quadruplex structure split from the octaplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*AP*GP*AP*GP*C)-3', POTASSIUM ION | | Authors: | Kondo, J, Umeda, S, Sunami, T, Takenaka, A. | | Deposit date: | 2003-11-03 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats
Nucleic Acids Res., 32, 2004
|
|
1V3P
 
 | | Crystal structure of d(GCGAGAGC): the DNA octaplex structure with I-motif of G-quartet | | Descriptor: | 5'-D(*GP*(C38)P*GP*AP*GP*AP*GP*C)-3', POTASSIUM ION | | Authors: | Kondo, J, Umeda, S, Sunami, T, Takenaka, A. | | Deposit date: | 2003-11-03 | | Release date: | 2004-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats
Nucleic Acids Res., 32, 2004
|
|
1ZYG
 
 | | Structure of a Supercoiling Responsive DNA Site | | Descriptor: | 5'-D(*CP*AP*AP*CP*CP*CP*GP*GP*GP*TP*TP*G)-3' | | Authors: | Bae, S.H, Yun, S.H, Sun, D, Lim, H.M, Choi, B.S. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic basis of a supercoiling-responsive DNA element
Nucleic Acids Res., 34, 2006
|
|
1ZYF
 
 | | Structure of a Supercoiling Responsive DNA Site | | Descriptor: | 5'-D(*CP*AP*AP*CP*CP*AP*TP*GP*GP*TP*TP*G)-3' | | Authors: | Bae, S.H, Yun, S.H, Sun, D, Lim, H.M, Choi, B.S. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic basis of a supercoiling-responsive DNA element
Nucleic Acids Res., 34, 2006
|
|
1ZYH
 
 | | Structure of a Supercoiling Responsive DNA site | | Descriptor: | 5'-D(*CP*AP*AP*CP*CP*AP*GP*GP*GP*TP*TP*G)-3', 5'-D(*CP*AP*AP*CP*CP*CP*TP*GP*GP*TP*TP*G)-3' | | Authors: | Bae, S.H, Yun, S.H, Sun, D, Lim, H.M, Choi, B.S. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic basis of a supercoiling-responsive DNA element
Nucleic Acids Res., 34, 2006
|
|
3TTZ
 
 | | Crystal structure of a topoisomerase ATPase inhibitor | | Descriptor: | 2-[(3S,4R)-4-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}-3-fluoropiperidin-1-yl]-1,3-thiazole-5-carboxylic acid, DNA gyrase subunit B, MAGNESIUM ION | | Authors: | Boriack-Sjodin, P.A, Read, J, Eakin, A.E, Sherer, B.A. | | Deposit date: | 2011-09-15 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Pyrrolamide DNA gyrase inhibitors: Optimization of antibacterial activity and efficacy.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1WUD
 
 | | E. coli RecQ HRDC domain | | Descriptor: | ATP-dependent DNA helicase recQ | | Authors: | Bernstein, D.A, Keck, J.L. | | Deposit date: | 2004-12-04 | | Release date: | 2005-08-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conferring Substrate Specificity to DNA Helicases: Role of the RecQ HRDC Domain
STRUCTURE, 13, 2005
|
|
3HTN
 
 | |
1TLF
 
 | |
3K0B
 
 | | Crystal structure of a predicted N6-adenine-specific DNA methylase from Listeria monocytogenes str. 4b F2365 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, predicted N6-adenine-specific DNA methylase | | Authors: | Nocek, B, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-24 | | Release date: | 2009-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a predicted N6-adenine-specific DNA methylase from Listeria monocytogenes str. 4b F2365
To be Published
|
|
1VPB
 
 | |
1XHV
 
 | | HincII bound to cleaved cognate DNA GTCGAC and Mn2+ | | Descriptor: | 5'-D(*GP*CP*CP*GP*GP*TP*C)-3', 5'-D(P*GP*AP*CP*CP*GP*G)-3', MANGANESE (II) ION, ... | | Authors: | Etzkorn, C, Horton, N.C. | | Deposit date: | 2004-09-20 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic Insights from the Structures of HincII Bound to Cognate DNA Cleaved from Addition of Mg(2+) and Mn(2+)
J.Mol.Biol., 343, 2004
|
|
1PUY
 
 | | 1.5 A resolution structure of a synthetic DNA hairpin with a stilbenediether linker | | Descriptor: | 5'-D(*GP*TP*TP*TP*TP*GP*(S02)P*CP*AP*AP*AP*AP*C)-3', MAGNESIUM ION | | Authors: | Egli, M, Tereshko, V, Murshudov, G, Sanishvili, R, Liu, X, Lewis, F.D. | | Deposit date: | 2003-06-25 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Face-to-face and edge-to-face pi-pi interactions in a synthetic DNA hairpin with a stilbenediether linker
J.Am.Chem.Soc., 125, 2003
|
|
1O0K
 
 | | Structure of the First Parallel DNA Quadruplex-drug Complex | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*T)-3', DAUNOMYCIN, SODIUM ION | | Authors: | Clark, G.R, Pytel, P.D, Squire, C.J, Neidle, S. | | Deposit date: | 2003-02-22 | | Release date: | 2003-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structure of the First Parallel DNA Quadruplex-drug Complex
J.Am.Chem.Soc., 125, 2003
|
|
3IUO
 
 | | The Crystal Structure of the C-terminal domain of the ATP-dependent DNA helicase RecQ from Porphyromonas gingivalis to 1.6A | | Descriptor: | ATP-dependent DNA helicase RecQ, CHLORIDE ION, SODIUM ION | | Authors: | Stein, A.J, Sather, A, Duggan, E, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-08 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the C-terminal domain of the ATP-dependent DNA helicase RecQ from Porphyromonas gingivalis to 1.6A
To be Published
|
|
1XHU
 
 | | HincII bound to cleaved, cognate DNA containing GTCGAC | | Descriptor: | 5'-D(*GP*CP*CP*GP*GP*TP*C)-3', 5'-D(P*GP*AP*CP*CP*GP*G)-3', Type II restriction enzyme HincII | | Authors: | Etzkorn, C, Horton, N.C. | | Deposit date: | 2004-09-20 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Mechanistic Insights from the Structures of HincII Bound to Cognate DNA Cleaved from Addition of Mg(2+) and Mn(2+)
J.Mol.Biol., 343, 2004
|
|
4B87
 
 | | Crystal structure of human DNA cross-link repair 1A | | Descriptor: | 1,2-ETHANEDIOL, DNA CROSS-LINK REPAIR 1A PROTEIN, ZINC ION | | Authors: | Allerston, C.K, Berridge, G, Carpenter, E.P, Kochan, G, Krojer, T, Mahajan, P, Vollmar, M, Yue, W.W, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2012-08-24 | | Release date: | 2012-11-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structure of Human DNA Cross-Link Repair 1A
To be Published
|
|
1X6W
 
 | |
