1FQC
 
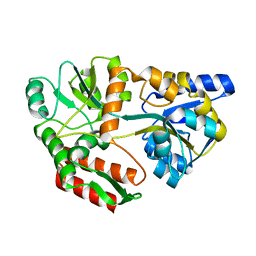 | | CRYSTAL STRUCTURE OF MALTOTRIOTOL BOUND TO CLOSED-FORM MALTODEXTRIN BINDING PROTEIN | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-D-glucose | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
3QS9
 
 | |
6TF0
 
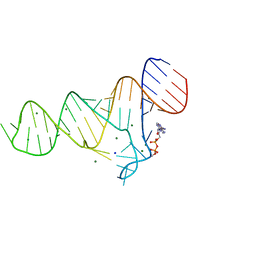 | | Crystal structure of the ADP-binding domain of the NAD+ riboswitch with Nicotinamide adenine dinucleotide, reduced (NADH) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Chains: A, MAGNESIUM ION, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2019-11-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and ligand binding of the ADP-binding domain of the NAD+ riboswitch.
Rna, 26, 2020
|
|
6TF3
 
 | |
6TFE
 
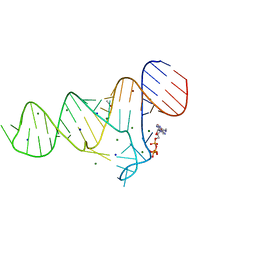 | |
4MSL
 
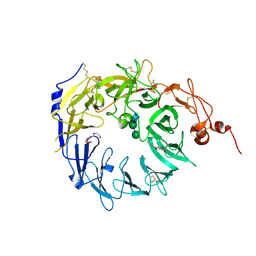 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with AF40431 | | Descriptor: | N-[(7-hydroxy-4-methyl-2-oxo-2H-chromen-8-yl)methyl]-L-leucine, Sortilin, TETRAETHYLENE GLYCOL, ... | | Authors: | Andersen, J.L, Strandbygaard, D, Thirup, S. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of the first small-molecule ligand of the neuronal receptor sortilin and structure determination of the receptor-ligand complex.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4A8G
 
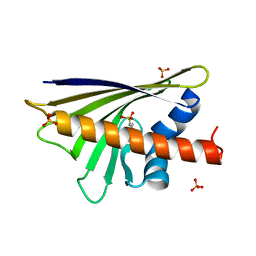 | |
4N33
 
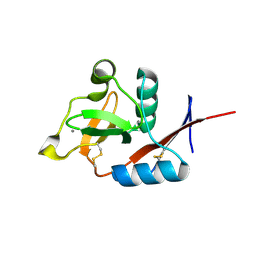 | | Structure of langerin CRD complexed with GlcNAc-beta1-3Gal-beta1-4Glc-beta-CH2CH2N3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, C-type lectin domain family 4 member K, ... | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
1RY7
 
 | | Crystal Structure of the 3 Ig form of FGFR3c in complex with FGF1 | | Descriptor: | Fibroblast growth factor receptor 3, Heparin-binding growth factor 1 | | Authors: | Olsen, S.K, Ibrahimi, O.A, Raucci, A, Zhang, F, Eliseenkova, A.V, Yayon, A, Basilico, C, Linhardt, R.J, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2003-12-19 | | Release date: | 2004-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
3U7U
 
 | | Crystal structure of extracellular region of human epidermal growth factor receptor 4 in complex with neuregulin-1 beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuregulin 1, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Liu, P, Cleveland IV, T.E, Bouyain, S, Longo, P.A, Leahy, D.J. | | Deposit date: | 2011-10-14 | | Release date: | 2012-08-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | A single ligand is sufficient to activate EGFR dimers.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NAW
 
 | | Crystal structure of E. coli O157:H7 effector protein NleL | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Lin, D.Y, Chen, J. | | Deposit date: | 2010-06-02 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and Structural Studies of a HECT-like Ubiquitin Ligase from Escherichia coli O157:H7.
J.Biol.Chem., 286, 2011
|
|
3K6V
 
 | | M. acetivorans Molybdate-Binding Protein (ModA) in Citrate-Bound Open Form | | Descriptor: | CITRIC ACID, Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1EPP
 
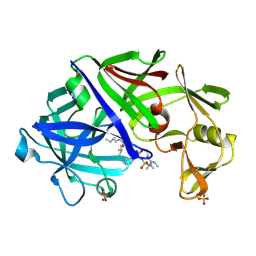 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-130,693 (MAS PHE LYS+MTF STA MBA) | | Descriptor: | ENDOTHIAPEPSIN, N-(dimethylsulfamoyl)-L-phenylalanyl-N-[(1S,2S)-2-hydroxy-4-{[(2S)-2-methylbutyl]amino}-1-(2-methylpropyl)-4-oxobutyl]-N~6~-(methylcarbamothioyl)-L-lysinamide, SULFATE ION | | Authors: | Wallace, B.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analyses of ligand binding in five endothiapepsin crystal complexes and their use in the design and evaluation of novel renin inhibitors.
J.Med.Chem., 36, 1993
|
|
1JDX
 
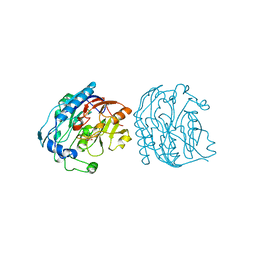 | |
5PZN
 
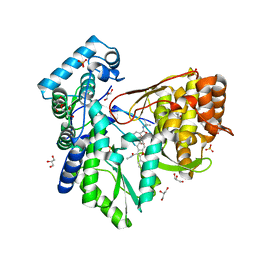 | |
2EUK
 
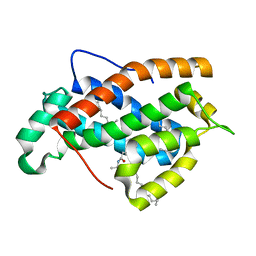 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with 24:1 Galactosylceramide | | Descriptor: | (15E)-TETRACOS-15-ENOIC ACID, Glycolipid transfer protein, N-OCTANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
3NB2
 
 | | Crystal structure of E. coli O157:H7 effector protein NleL | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Lin, D.Y, Chen, J. | | Deposit date: | 2010-06-02 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and Structural Studies of a HECT-like Ubiquitin Ligase from Escherichia coli O157:H7.
J.Biol.Chem., 286, 2011
|
|
2EVT
 
 | | Crystal structure of D48V mutant of human Glycolipid Transfer Protein | | Descriptor: | Glycolipid transfer protein, HEXANE | | Authors: | Malinina, L, Malakhova, M.L, Teplov, A, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
5PZO
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE C316N IN COMPLEX WITH 2-(4-FLUOROPHENYL)-N-METHYL-5-[3-({2-METHYL-1-OXO-1-[(1,3,4-THIADIAZOL-2-YL)AMINO]PROPAN-2-YL}CARBAMOYL)PHENYL]-1-BENZOFURAN-3-CARBOXAMIDE | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 2-(4-fluorophenyl)-N-methyl-5-[3-({2-methyl-1-oxo-1-[(1,3,4-thiadiazol-2-yl)amino]propan-2-yl}carbamoyl)phenyl]-1-benzofuran-3-carboxamide, GLYCEROL, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Hepatitis C Virus NS5B Replicase Palm Site Allosteric Inhibitor (BMS-929075) Advanced to Phase 1 Clinical Studies.
J. Med. Chem., 60, 2017
|
|
7FFP
 
 | | Crystal structure of di-peptidase-E from Xenopus laevis | | Descriptor: | ASPARTIC ACID, Alpha-aspartyl dipeptidase, CALCIUM ION | | Authors: | Kumar, A, Singh, R, Makde, R.D. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aspartyl dipeptidase from Xenopus laevis revealed ligand binding induced loop ordering and catalytic triad assembly.
Proteins, 90, 2022
|
|
3CKD
 
 | | Crystal structure of the C-terminal domain of the Shigella type III effector IpaH | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Invasion plasmid antigen, ... | | Authors: | Lam, R, Singer, A.U, Cuff, M.E, Skarina, T, Kagan, O, DiLeo, R, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1UZ0
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with Glc-4Glc-3Glc-4Glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
3K6X
 
 | | M. acetivorans Molybdate-Binding Protein (ModA) in Molybdate-Bound Close Form with 2 Molecules in Asymmetric Unit Forming Beta Barrel | | Descriptor: | MOLYBDATE ION, SULFATE ION, Solute-binding protein MA_0280 | | Authors: | Chan, S, Chernishof, I, Giuroiu, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3WHB
 
 | |
3CNV
 
 | | Crystal structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica | | Descriptor: | CHLORIDE ION, CITRATE ANION, Putative GntR-family transcriptional regulator | | Authors: | Zimmerman, M.D, Xu, X, Cui, H, Filippova, E.V, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-26 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica.
To be Published
|
|
