2N2V
 
 | |
1A1K
 
 | | RADR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GACC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*TP*CP*CP*CP*AP*CP*GP*C)-3'), RADR ZIF268 VARIANT, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
2NNH
 
 | | CYP2C8dH complexed with 2 molecules of 9-cis retinoic acid | | Descriptor: | (9cis)-retinoic acid, Cytochrome P450 2C8, PALMITIC ACID, ... | | Authors: | Schoch, G.A, Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2006-10-24 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid.
J.Biol.Chem., 283, 2008
|
|
1AHD
 
 | | DETERMINATION OF THE NMR SOLUTION STRUCTURE OF AN ANTENNAPEDIA HOMEODOMAIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3'), Homeotic protein antennapedia | | Authors: | Billeter, M, Qian, Y.Q, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1993-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex.
J.Mol.Biol., 234, 1993
|
|
2NO3
 
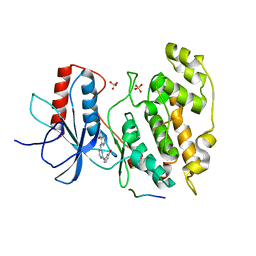 | | Novel 4-anilinopyrimidines as potent JNK1 Inhibitors | | Descriptor: | 2-({2-[(3-HYDROXYPHENYL)AMINO]PYRIMIDIN-4-YL}AMINO)BENZAMIDE, C-JUN-AMINO-TERMINAL KINASE-INTERACTING protein 1, Mitogen-activated protein kinase 8, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-10-24 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of a new class of 4-anilinopyrimidines as potent c-Jun N-terminal kinase inhibitors: Synthesis and SAR studies.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3MA6
 
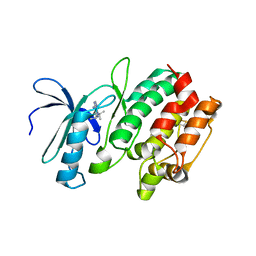 | | Crystal structure of kinase domain of TgCDPK1 in presence of 3BrB-PP1 | | Descriptor: | 3-(3-bromobenzyl)-1-tert-butyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1 | | Authors: | Wernimont, A.K, Qiu, W, Amani, M, Artz, J.D, Hassani, A.A, Senisterra, G, Vedadi, M, Sibley, L.D, Lourido, S, Shokat, K, Zhang, C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-07-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of kinase domain of TgCDPK1 in presence of 3BrB-PP1
To be Published
|
|
2N42
 
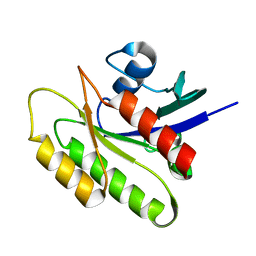 | | EC-NMR Structure of Human H-RasT35S mutant protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data | | Descriptor: | GTPase HRas | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
3MDF
 
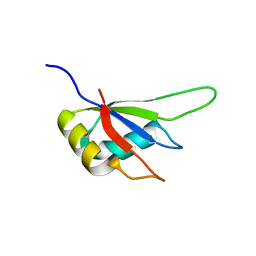 | | Crystal structure of the RRM domain of Cyclophilin 33 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Hom, R.A, Chang, P.Y, Roy, S, Mussleman, C.A, Glass, K.C, Seleznevia, A.I, Gozani, O, Ismagilov, R.F, Cleary, M.L, Kutateladze, T.G. | | Deposit date: | 2010-03-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular mechanism of MLL PHD3 and RNA recognition by the Cyp33 RRM domain.
J.Mol.Biol., 400, 2010
|
|
1ANE
 
 | | ANIONIC TRYPSIN WILD TYPE | | Descriptor: | ANIONIC TRYPSIN, BENZAMIDINE | | Authors: | Fletterick, R.J, Mcgrath, M.E. | | Deposit date: | 1994-12-21 | | Release date: | 1997-04-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an engineered, metal-actuated switch in trypsin.
Biochemistry, 32, 1993
|
|
3MDM
 
 | | Thioperamide complex of Cytochrome P450 46A1 | | Descriptor: | Cholesterol 24-hydroxylase, N-cyclohexyl-4-(1H-imidazol-5-yl)piperidine-1-carbothioamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mast, N, Charvet, C, Pikuleva, I, Stout, C.D. | | Deposit date: | 2010-03-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain.
J.Biol.Chem., 285, 2010
|
|
3MB6
 
 | | Human CK2 catalytic domain in complex with a difurane derivative inhibitor (CPA) | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, naphtho[2,1-b:7,6-b']difuran-2,8-dicarboxylic acid | | Authors: | Reiser, J.-B, Prudent, R, Claude, C. | | Deposit date: | 2010-03-25 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New potent dual inhibitors of CK2 and Pim kinases: discovery and structural insights.
Faseb J., 24, 2010
|
|
1A94
 
 | | STRUCTURAL BASIS FOR SPECIFICITY OF RETROVIRAL PROTEASES | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE | | Authors: | Wu, J, Adomat, J.M, Ridky, T.W, Louis, J.M, Leis, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 1998-04-16 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for specificity of retroviral proteases.
Biochemistry, 37, 1998
|
|
2MS1
 
 | |
1A00
 
 | | HEMOGLOBIN (VAL BETA1 MET, TRP BETA37 TYR) MUTANT | | Descriptor: | HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Arnone, A. | | Deposit date: | 1997-12-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structures of human hemoglobin with mutations at tryptophan 37beta: structural basis for a high-affinity T-state,.
Biochemistry, 37, 1998
|
|
1A1B
 
 | |
1A1L
 
 | | ZIF268 ZINC FINGER-DNA COMPLEX (GCAC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*CP*CP*AP*CP*GP*C)-3'), PROTEIN (ZIF268 ZINC FINGER PEPTIDE), ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1AFA
 
 | |
2N3B
 
 | | Structure of oxidized horse heart cytochrome c encapsulated in reverse micelles | | Descriptor: | Cytochrome c, HEME C | | Authors: | O'Brien, E.S, Nucci, N.V, Fuglestad, B, Tommos, C, Wand, A. | | Deposit date: | 2015-05-27 | | Release date: | 2015-10-28 | | Last modified: | 2016-02-03 | | Method: | SOLUTION NMR | | Cite: | Defining the Apoptotic Trigger: THE INTERACTION OF CYTOCHROME c AND CARDIOLIPIN.
J.Biol.Chem., 290, 2015
|
|
1A5P
 
 | | C[40,95]A VARIANT OF BOVINE PANCREATIC RIBONUCLEASE A | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Pearson, M.A, Karplus, P.A, Dodge, R.W, Laity, J.H, Scheraga, H.A. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of two mutants that have implications for the folding of bovine pancreatic ribonuclease A.
Protein Sci., 7, 1998
|
|
1AN4
 
 | | STRUCTURE AND FUNCTION OF THE B/HLH/Z DOMAIN OF USF | | Descriptor: | DNA (5'-D(*CP*AP*CP*CP*CP*GP*GP*TP*CP*AP*CP*GP*TP*GP*GP*CP*C P*TP*AP*CP*A)-3'), DNA (5'-D(*GP*TP*GP*TP*AP*GP*GP*CP*CP*AP*CP*GP*TP*GP*AP*CP*C P*GP*GP*GP*T)-3'), PROTEIN (UPSTREAM STIMULATORY FACTOR) | | Authors: | Ferre-D'Amare, A.R, Pognonec, P, Roeder, R.G, Burley, S.K. | | Deposit date: | 1997-03-15 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and function of the b/HLH/Z domain of USF.
EMBO J., 13, 1994
|
|
2MRK
 
 | | Fyn SH2 domain in complex with the natural inhibitory phosphotyrosine peptide | | Descriptor: | C-terminal Tyrosine-protein kinase Fyn, Tyrosine-protein kinase Fyn | | Authors: | Huculeci, R, Buts, L, Lenaerts, A.J, van Nuland, N. | | Deposit date: | 2014-07-09 | | Release date: | 2015-07-15 | | Last modified: | 2016-12-14 | | Method: | SOLUTION NMR | | Cite: | Dynamically Coupled Residues within the SH2 Domain of FYN Are Key to Unlocking Its Activity.
Structure, 24, 2016
|
|
7D6L
 
 | |
2MQV
 
 | |
1AJE
 
 | | CDC42 FROM HUMAN, NMR, 20 STRUCTURES | | Descriptor: | CDC42HS | | Authors: | Feltham, J.L, Dotsch, V, Raza, S, Manor, D, Cerione, R.A, Sutcliffe, M.J, Wagner, G, Oswald, R.E. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Definition of the switch surface in the solution structure of Cdc42Hs.
Biochemistry, 36, 1997
|
|
1ABY
 
 | | CYANOMET RHB1.1 (RECOMBINANT HEMOGLOBIN) | | Descriptor: | CYANIDE ION, HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kundrot, C.E, Kroeger, K.S. | | Deposit date: | 1997-01-29 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of a hemoglobin-based blood substitute: insights into the function of allosteric proteins.
Structure, 5, 1997
|
|
