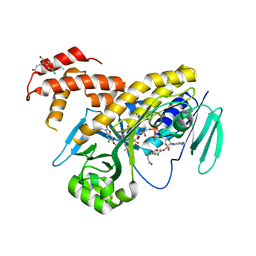
5MZC
 
 | |
8RTH
 
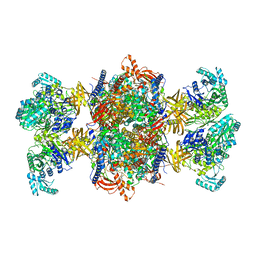 | | Trypanosoma brucei 3-methylcrotonyl-CoA carboxylase | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, putative, 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ... | | Authors: | Ruiz, F.M, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2024-01-26 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The cryo-EM structure of trypanosome 3-methylcrotonyl-CoA carboxylase provides mechanistic and dynamic insights into its enzymatic function.
Structure, 32, 2024
|
|
5MZI
 
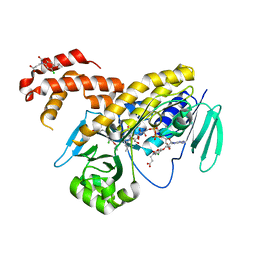 | | Pseudomonas fluorescens kynurenine 3-monooxygenase (KMO) in complex with 3-(5-chloro-6-cyclopropoxy-2-oxo-2,3-dihydro-1,3-benzoxazol-3-yl)propanoic acid | | Descriptor: | 3-(5-chloro-6-cyclopropoxy-2-oxo-2,3-dihydro-1,3-benzoxazol-3-yl)propanoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rowland, P. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Development of a Series of Kynurenine 3-Monooxygenase Inhibitors Leading to a Clinical Candidate for the Treatment of Acute Pancreatitis.
J. Med. Chem., 60, 2017
|
|
5N7T
 
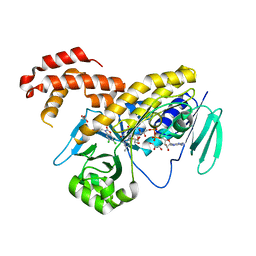 | | Pseudomonas fluorescens kynurenine 3-monooxygenase (KMO) in complex with 3-(5,6-dichloro-2-oxo-2,3-dihydro-1,3-benzoxazol-3-yl)propanoic acid | | Descriptor: | 3-(5,6-DICHLORO-2-OXOBENZO[D]OXAZOL-3(2H)-YL)PROPANOIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rowland, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The discovery of potent and selective kynurenine 3-monooxygenase inhibitors for the treatment of acute pancreatitis.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
2CIO
 
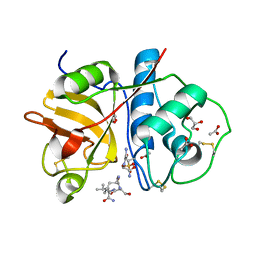 | |
5NAB
 
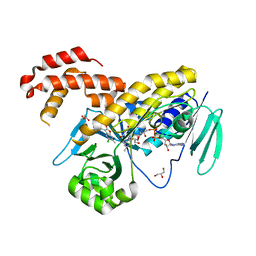 | |
4OL9
 
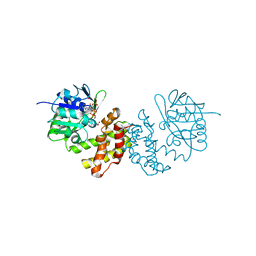 | |
5GNX
 
 | | The E171Q mutant structure of Bgl6 | | Descriptor: | Beta-glucosidase, GLYCEROL, PROPANOIC ACID, ... | | Authors: | Xie, W, Pang, P, Cao, L.C, Liu, Y.H, Wang, Z. | | Deposit date: | 2016-07-25 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism.
J. Struct. Biol., 198, 2017
|
|
4MSS
 
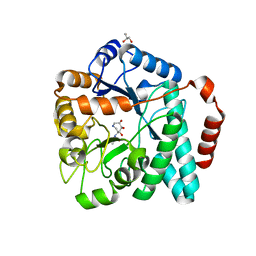 | | Crystal structure of Burkholderia cenocepacia family 3 glycoside hydrolase (NagZ) bound to (3S,4R,5R,6S)-3-acetamido-4,5,6-trihydroxyazepane | | Descriptor: | Beta-hexosaminidase 1, GLYCEROL, N-[(3S,4R,5R,6S)-4,5,6-trihydroxyazepan-3-yl]acetamide | | Authors: | Vadlamani, G, Mark, B.L. | | Deposit date: | 2013-09-18 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective trihydroxyazepane NagZ inhibitors increase sensitivity of Pseudomonas aeruginosa to beta-lactams.
Chem.Commun.(Camb.), 49, 2013
|
|
5EIZ
 
 | |
5JV5
 
 | |
5KAP
 
 | |
6BG7
 
 | |
6BG6
 
 | |
5K29
 
 | | Trypanosoma brucei bromodomain BDF5 (Tb427tmp.01.5000) | | Descriptor: | UNKNOWN ATOM OR ION, uncharacterized protein BDF5 | | Authors: | Lin, Y.H, Tempel, W, Walker, J.R, Loppnau, P, Amani, M, Hou, C.F.D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-18 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trypanosoma brucei bromodomain BDF5 (Tb427tmp.01.5000)
To Be Published
|
|
4QP3
 
 | |
7T5J
 
 | | Crystal Structure of Strictosidine Synthase in complex with S-IPA (2-(1H-indol-3-yl) propan-1-amine) | | Descriptor: | (2S)-2-(1H-indol-3-yl)propan-1-amine, Strictosidine synthase | | Authors: | Liu, H, Panjikar, S, Futamura, Y, Shao, N, Osada, H, Zou, H. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | beta-Branched Tryptamine Provoked Non Sandwich-Like-Mode Catalysis of Strictosidine Synthase to Antimalarial Indole Alkaloids
to be published
|
|
7T5I
 
 | | Crystal Structure of Strictosidine Synthase in complex with R-IPA(2-(1H-indol-3-yl) propan-1-amine) | | Descriptor: | (2R)-2-(1H-indol-3-yl)propan-1-amine, Strictosidine synthase | | Authors: | Liu, H, Panjikar, S, Futamura, Y, Shao, N, Osada, H, Zou, H. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Beta-Branched Tryptamine Provoked Non Sandwich-Like-Mode Catalysis of Strictosidine Synthase to Antimalarial Indole Alkaloids
to be published
|
|
9AXO
 
 | |
6R9P
 
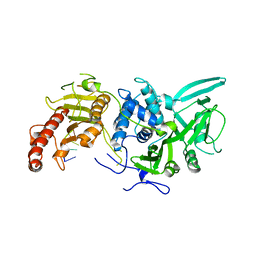 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with AAUUAA RNA | | Descriptor: | AAUUAA RNA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | Authors: | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6R9O
 
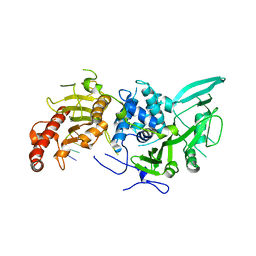 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with AAGGA RNA | | Descriptor: | AAGGA RNA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | Authors: | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.319 Å) | | Cite: | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6XOC
 
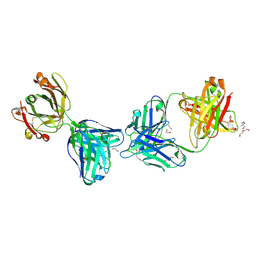 | | Crystal structure of glVRC01 Fab in complex with anti-idiotypic iv4 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Aljedani, S.S, Weidle, C, Pancera, M. | | Deposit date: | 2020-07-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development of a VRC01-class germline targeting immunogen derived from anti-idiotypic antibodies.
Cell Rep, 35, 2021
|
|
6R9M
 
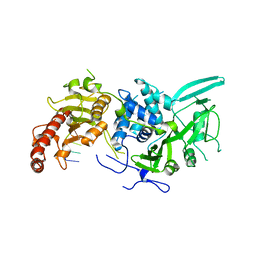 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with AAGGAA RNA | | Descriptor: | AAGGAA RNA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | Authors: | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.329 Å) | | Cite: | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6R9Q
 
 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with AACCAA RNA | | Descriptor: | AACCAA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | Authors: | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.079 Å) | | Cite: | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6R9J
 
 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with A7 RNA | | Descriptor: | A7 RNA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | Authors: | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.326 Å) | | Cite: | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
