8ULG
 
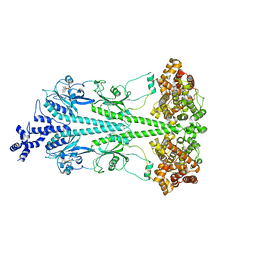 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8UFI
 
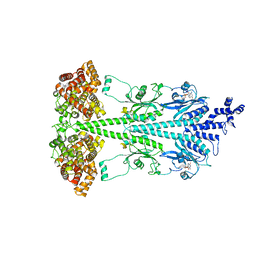 | | Cryo-EM structure of bovine phosphodiesterase 6 | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8UGB
 
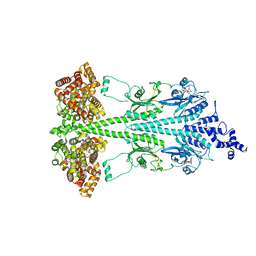 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to udenafil | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-05 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8UGS
 
 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
2YAK
 
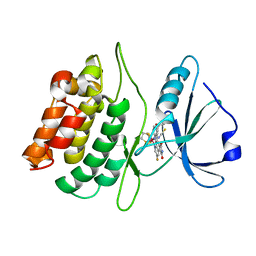 | | Structure of death-associated protein Kinase 1 (dapk1) in complex with a ruthenium octasporine ligand (OSV) | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 1, RUTHENIUM OCTASPORINE 4 | | Authors: | Feng, L, Geisselbrecht, Y, Blanck, S, Wilbuer, A, Atilla-Gokcumen, G.E, Filippakopoulos, P, Kraeling, K, Celik, M.A, Harms, K, Maksimoska, J, Marmorstein, R, Frenking, G, Knapp, S, Essen, L.-O, Meggers, E. | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structurally Sophisticated Octahedral Metal Complexes as Highly Selective Protein Kinase Inhibitors.
J.Am.Chem.Soc., 133, 2011
|
|
4NDD
 
 | | X-ray structure of a double mutant of calexcitin - a neuronal calcium-signalling protein | | Descriptor: | CALCIUM ION, Calexcitin | | Authors: | Erskine, P.T, Fokas, A, Muriithi, C, Razzall, E, Bowyer, A, Findlow, I.S, Werner, J.M, Wallace, B.A, Wood, S.P, Cooper, J.B. | | Deposit date: | 2013-10-25 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray, spectroscopic and normal-mode dynamics of calexcitin: structure-function studies of a neuronal calcium-signalling protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4NDB
 
 | | X-ray structure of a mutant (T61D) of calexcitin - a neuronal calcium-signalling protein | | Descriptor: | CALCIUM ION, Calexcitin | | Authors: | Erskine, P.T, Fokas, A, Muriithi, C, Razzall, E, Bowyer, A, Findlow, I.S, Werner, J.M, Wallace, B.A, Wood, S.P, Cooper, J.B. | | Deposit date: | 2013-10-25 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray, spectroscopic and normal-mode dynamics of calexcitin: structure-function studies of a neuronal calcium-signalling protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4NDC
 
 | | X-ray structure of a mutant (T188D) of calexcitin - a neuronal calcium-signalling protein | | Descriptor: | CALCIUM ION, Calexcitin | | Authors: | Erskine, P.T, Fokas, A, Muriithi, C, Razzall, E, Bowyer, A, Findlow, I.S, Werner, J.M, Wallace, B.A, Wood, S.P, Cooper, J.B. | | Deposit date: | 2013-10-25 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray, spectroscopic and normal-mode dynamics of calexcitin: structure-function studies of a neuronal calcium-signalling protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1PHK
 
 | | TWO STRUCTURES OF THE CATALYTIC DOMAIN OF PHOSPHORYLASE, KINASE: AN ACTIVE PROTEIN KINASE COMPLEXED WITH NUCLEOTIDE, SUBSTRATE-ANALOGUE AND PRODUCT | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PHOSPHORYLASE KINASE | | Authors: | Owen, D.J, Noble, M.E.M, Garman, E.F, Papageorgiou, A.C, Johnson, L.N. | | Deposit date: | 1996-03-15 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two structures of the catalytic domain of phosphorylase kinase: an active protein kinase complexed with substrate analogue and product.
Structure, 3, 1995
|
|
1QL6
 
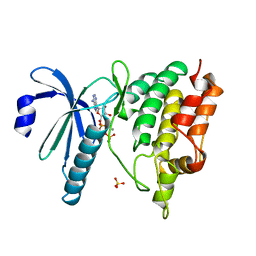 | | THE CATALYTIC MECHANISM OF PHOSPHORYLASE KINASE PROBED BY MUTATIONAL STUDIES | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PHOSPHORYLASE KINASE, ... | | Authors: | Skamnaki, V.T, Owen, D.J, Noble, M.E.M, Lowe, E.D, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1999-08-24 | | Release date: | 1999-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic Mechanism of Phosphorylase Kinase Probed by Mutational Studies.
Biochemistry, 38, 1999
|
|
1TLK
 
 | |
5WST
 
 | | Crystal structure of Myo7a SAH | | Descriptor: | Unconventional myosin-VIIa | | Authors: | Li, J, Chen, Y, Deng, Y, Lu, Q, Zhang, M. | | Deposit date: | 2016-12-08 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension
Structure, 25, 2017
|
|
4V24
 
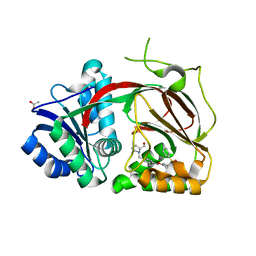 | | Sphingosine kinase 1 in complex with PF-543 | | Descriptor: | ACETATE ION, SPHINGOSINE KINASE 1, {(2R)-1-[4-({3-METHYL-5-[(PHENYLSULFONYL)METHYL]PHENOXY}METHYL)BENZYL]PYRROLIDIN-2-YL}METHANOL | | Authors: | Elkins, J.M, Wang, J, Sorrell, F, Tallant, C, Wang, D, Shrestha, L, Bountra, C, von Delft, F, Knapp, S, Edwards, A. | | Deposit date: | 2014-10-05 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Sphingosine Kinase 1 with Pf-543.
Acs Med.Chem.Lett., 5, 2014
|
|
8Q6T
 
 | |
1UGJ
 
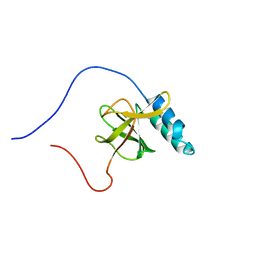 | | Solution structure of a murine hypothetical protein from RIKEN cDNA 2310057J16 | | Descriptor: | RIKEN cDNA 2310057J16 protein | | Authors: | Nagashima, T, Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-16 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a murine hypothetical protein from RIKEN cDNA 2310057J16
To be Published
|
|
7D2K
 
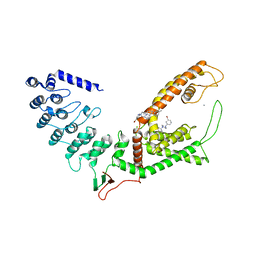 | | Crystal structure of rat TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor Br-cis-22a | | Descriptor: | 1-(5-bromanylpyridin-3-yl)-4-[4-(3-methylphenyl)cyclohexyl]piperazin-4-ium, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Singh, A.K, Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.698 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7U0T
 
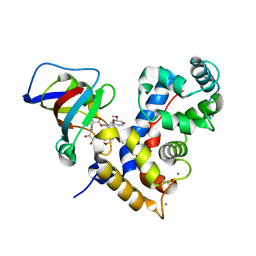 | | Crystal Structure of a human Calcineurin A - Calcineurin B fusion bound to FKBP12 and FK-520 | | Descriptor: | (3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,22R,26aS)-8-ethyl-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-5,6,8,11,12,13,14,15,16,17,18,19,24,25,26,26a-hexadecahydro-3H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosine-1,7,20,21(4H,23H)-tetrone, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Fox III, D, Mayclin, S.J, DeBouver, N.D, Hoy, M.J, Heitman, J, Lorimer, D.D, Horanyi, P.S, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus.
Mbio, 13, 2022
|
|
1E7O
 
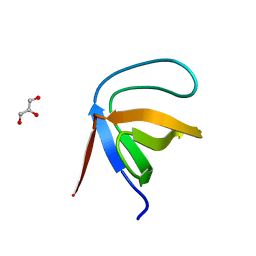 | | A-SPECTRIN SH3 DOMAIN A11V, V23L, M25V, V44I, V58L MUTATIONS | | Descriptor: | GLYCEROL, SPECTRIN ALPHA CHAIN | | Authors: | Vega, M.C, Serrano, L. | | Deposit date: | 2000-08-31 | | Release date: | 2003-05-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Thermodynamic and Kinetic Analysis of the Folding Pathway of an SH3 Domain Entropically Stabilised by a Redesigned Hydrophobic Core
J.Mol.Biol., 328, 2003
|
|
5VSN
 
 | |
5X2D
 
 | |
2NYJ
 
 | | Crystal structure of the ankyrin repeat domain of TRPV1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Jin, X, Gaudet, R. | | Deposit date: | 2006-11-20 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Ankyrin Repeats of TRPV1 Bind Multiple Ligands and Modulate Channel Sensitivity.
Neuron, 54, 2007
|
|
3BJC
 
 | | Crystal structure of the PDE5A catalytic domain in complex with a novel inhibitor | | Descriptor: | 5-ethoxy-4-(1-methyl-7-oxo-3-propyl-6,7-dihydro-1H-pyrazolo[4,3-d]pyrimidin-5-yl)thiophene-2-sulfonamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, G, Wang, H, Howard, R, Cai, J, Wan, Y, Ke, H. | | Deposit date: | 2007-12-03 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An insight into the pharmacophores of phosphodiesterase-5 inhibitors from synthetic and crystal structural studies
BIOCHEM.PHARM., 75, 2008
|
|
7LXC
 
 | |
8IE6
 
 | | Crystal structure of DAPK1 in complex with pinostilbene | | Descriptor: | 3-[(E)-2-(4-hydroxyphenyl)ethenyl]-5-methoxy-phenol, Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Characterization of the molecular interactions between resveratrol derivatives and death-associated protein kinase 1.
Febs J., 290, 2023
|
|
8IE5
 
 | | Crystal structure of DAPK1 in complex with oxyresveratrol | | Descriptor: | Death-associated protein kinase 1, SULFATE ION, trans-oxyresveratrol | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Characterization of the molecular interactions between resveratrol derivatives and death-associated protein kinase 1.
Febs J., 290, 2023
|
|
