7ZH4
 
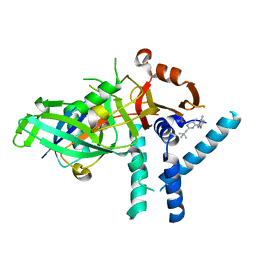 | | USP1 bound to ML323 and ubiquitin conjugated to FANCD2 (focused refinement) | | Descriptor: | 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phenyl]methyl]pyrimidin-4-amine, Ubiquitin carboxyl-terminal hydrolase 1, Ubiquitin-60S ribosomal protein L40, ... | | Authors: | Rennie, M.L, Walden, H. | | Deposit date: | 2022-04-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site.
Sci Adv, 8, 2022
|
|
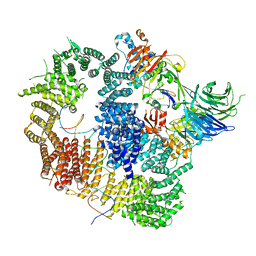
8A9J
 
 | |
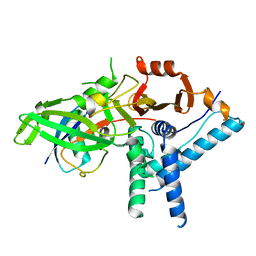
7ZH3
 
 | |
8C13
 
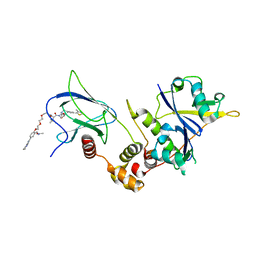 | | Crystal structure of pVHL:ElonginC:ElonginB complex bound to PROTAC JW48 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[3-[2-[2-[2-(acetamidomethyl)-4-(6,7-dihydro-5~{H}-pyrrolo[1,2-a]imidazol-2-yl)phenoxy]ethoxy]ethoxy]propanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kraemer, A, Weckesser, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
8VLB
 
 | | Crystal structure of EloBC-VHL-CDO1 complex bound to compound 4 molecular glue | | Descriptor: | CITRIC ACID, Cysteine dioxygenase type 1, Elongin-B, ... | | Authors: | Shu, W, Ma, X, Tutter, A, Buckley, D, Golosov, A, Michaud, G. | | Deposit date: | 2024-01-11 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A small molecule VHL molecular glue degrader for cysteine dioxygenase 1
To Be Published
|
|
8P0F
 
 | | Crystal structure of the VCB complex with compound 1. | | Descriptor: | (3~{R},5~{R})-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-5-oxidanyl-2-oxidanylidene-1-pyridin-2-yl-piperidine-3-carboxamide, CHLORIDE ION, Elongin-B, ... | | Authors: | Bader, G, Boettcher, J, Wolkerstorfer, B. | | Deposit date: | 2023-05-10 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Drugit: Crowd-sourcing molecular design of non-peptidic VHL binders
Chemrxiv, 2023
|
|
8WQF
 
 | | cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CUX1 (conformation 2) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQE
 
 | | Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CUX1 (conformation 1) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQC
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CDK5R1 | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQG
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
2KWU
 
 | | Solution Structure of UBM2 of murine Polymerase iota in Complex with Ubiquitin | | Descriptor: | DNA polymerase iota, Ubiquitin | | Authors: | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | Deposit date: | 2010-04-19 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
2KWV
 
 | | Solution Structure of UBM1 of murine Polymerase iota in Complex with Ubiquitin | | Descriptor: | DNA polymerase iota, Ubiquitin | | Authors: | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | Deposit date: | 2010-04-20 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
2L0T
 
 | | Solution structure of the complex of ubiquitin and the VHS domain of Stam2 | | Descriptor: | Signal transducing adapter molecule 2, Ubiquitin | | Authors: | Lange, A, Hoeller, D, Wienk, H, Marcillat, O, Lancelin, J, Walker, O. | | Deposit date: | 2010-07-15 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR reveals a different mode of binding of the Stam2 VHS domain to ubiquitin and diubiquitin.
Biochemistry, 50, 2011
|
|
2LVO
 
 | | Structure of the gp78CUE domain bound to monubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LVP
 
 | | gp78CUE domain bound to the distal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2MBB
 
 | | Solution Structure of the human Polymerase iota UBM1-Ubiquitin Complex | | Descriptor: | Immunoglobulin G-binding protein G/DNA polymerase iota fusion protein, Polyubiquitin-B | | Authors: | Wang, S, Zhou, P. | | Deposit date: | 2013-07-29 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sparsely-sampled, high-resolution 4-D omit spectra for detection and assignment of intermolecular NOEs of protein complexes.
J.Biomol.Nmr, 59, 2014
|
|
2MBH
 
 | | NMR structure of EKLF(22-40)/Ubiquitin Complex | | Descriptor: | Krueppel-like factor 1, Ubiquitin | | Authors: | Raiola, L, Omichinski, J.G. | | Deposit date: | 2013-07-31 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a Noncovalent Complex between Ubiquitin and the Transactivation Domain of the Erythroid-Specific Factor EKLF.
Structure, 21, 2013
|
|
2LVQ
 
 | | gp78CUE domain bound to the proximal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2MCN
 
 | | Distinct ubiquitin binding modes exhibited by SH3 domains: molecular determinants and functional implications | | Descriptor: | CD2-associated protein, Ubiquitin | | Authors: | Ortega-Roldan, J, Salmon, L, Azuaga, A, Blackledge, M, Van Nuland, N. | | Deposit date: | 2013-08-22 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Distinct ubiquitin binding modes exhibited by SH3 domains: molecular determinants and functional implications.
Plos One, 8, 2013
|
|
2N3W
 
 | |
2N3U
 
 | |
2N9P
 
 | | Solution structure of RNF126 N-terminal zinc finger domain in complex with BAG6 Ubiquitin-like domain | | Descriptor: | E3 ubiquitin-protein ligase RNF126, Large proline-rich protein BAG6, ZINC ION | | Authors: | Martinez-Lumbreras, S, Krysztofinska, E.M, Thapaliya, A, Isaacson, R.L. | | Deposit date: | 2015-12-01 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into the E3 ligase, RNF126.
Sci Rep, 6, 2016
|
|
2NBW
 
 | | Solution structure of the Rpn1 T1 site with the Rad23 UBL domain | | Descriptor: | 26S proteasome regulatory subunit RPN1, UV excision repair protein RAD23 | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2016-03-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome.
Structure, 24, 2016
|
|
2NBV
 
 | |
2N3V
 
 | |
