2Q9M
 
 | | 4-Substituted Trinems as Broad Spectrum-Lactamase Inhibitors: Structure-based Design, Synthesis and Biological Activity | | Descriptor: | (1R,4S,7AS)-1-(1-FORMYLPROP-1-EN-1-YL)-4-METHOXY-2,4,5,6,7,7A-HEXAHYDRO-1H-ISOINDOLE-3-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Plantan, I, Selic, L, Mesar, T, Stefanic Anderluh, P, Oblak, M, Prezelj, A, Hesse, L, Andrejasic, M, Vilar, M, Turk, D, Kocijan, A, Prevec, T, Vilfan, G, Kocjan, D, Copar, A, Urleb, U, Solmajer, T. | | Deposit date: | 2007-06-13 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 4-Substituted Trinems as Broad Spectrum beta-Lactamase Inhibitors: Structure-Based Design, Synthesis, and Biological Activity
J.Med.Chem., 50, 2007
|
|
2PU4
 
 | |
2PU2
 
 | | AmpC beta-lactamase with bound Phthalamide inhibitor | | Descriptor: | 2-[(1R)-1-CARBOXY-2-(4-HYDROXYPHENYL)ETHYL]-1,3-DIOXOISOINDOLINE-5-CARBOXYLIC ACID, Beta-lactamase, PHOSPHATE ION | | Authors: | Babaoglu, K, Shoichet, B.K. | | Deposit date: | 2007-05-08 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase.
J.Med.Chem., 51, 2008
|
|
2P9V
 
 | | Structure of AmpC beta-lactamase with cross-linked active site after exposure to small molecule inhibitor | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Babaoglu, K, Wyrembak, P.N, Pelto, R.B, Shoichet, B.K, Pratt, R.F. | | Deposit date: | 2007-03-26 | | Release date: | 2007-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | O-Aryloxycarbonyl Hydroxamates: New beta-Lactamase Inhibitors That Cross-Link the Active Site.
J.Am.Chem.Soc., 129, 2007
|
|
2P74
 
 | |
2OV5
 
 | | Crystal structure of the KPC-2 carbapenemase | | Descriptor: | BICINE, Carbapenemase | | Authors: | Ke, W, Bethel, C.R, Thomson, J.M, Bonomo, R.A, van den Akker, F. | | Deposit date: | 2007-02-13 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of KPC-2: Insights into Carbapenemase Activity in Class A beta-Lactamases.
Biochemistry, 46, 2007
|
|
2I72
 
 | | AmpC beta-lactamase in complex with 5-diformylaminomethyl-benzo[b]thiophen-2-boronic acid | | Descriptor: | Beta-lactamase, {5-[(DIFORMYLAMINO)METHYL]-1-BENZOTHIEN-2-YL}BORONIC ACID | | Authors: | Venturelli, A, Cancian, L, Tondi, D, Morandi, F, Cannazza, G, Segatore, B, Prati, F, Amicosante, G, Shoichet, B.K, Costi, M.P. | | Deposit date: | 2006-08-30 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimizing Cell Permeation of an Antibiotic Resistance Inhibitor for Improved Efficacy
J.Med.Chem., 50, 2007
|
|
2HDU
 
 | |
2HDS
 
 | |
2HDR
 
 | |
2HDQ
 
 | |
2H5S
 
 | | SA2-13 penam sulfone complexed to wt SHV-1 beta-lactamase | | Descriptor: | (3R)-4-[(4-CARBOXYBUTANOYL)OXY]-N-[(1E)-3-OXOPROP-1-EN-1-YL]-3-SULFINO-D-VALINE, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, SHV-1 beta-lactamase | | Authors: | van den Akker, F, Padayatti, P.S. | | Deposit date: | 2006-05-27 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Rational Design of a beta-Lactamase Inhibitor Achieved via Stabilization of the trans-Enamine Intermediate: 1.28 A Crystal Structure of wt SHV-1 Complex with a Penam Sulfone.
J.Am.Chem.Soc., 128, 2006
|
|
2H10
 
 | |
2H0Y
 
 | |
2H0T
 
 | |
2GDN
 
 | |
2G2W
 
 | | Crystal Structure of the SHV D104K Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Reynolds, K.A, Thomson, J.M, Corbett, K.D, Bethel, C.R, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2006-02-16 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Computational Characterization of the SHV-1 beta-Lactamase-beta-Lactamase Inhibitor Protein Interface.
J.Biol.Chem., 281, 2006
|
|
2G2U
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Reynolds, K.A, Thomson, J.M, Corbett, K.D, Bethel, C.R, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2006-02-16 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Computational Characterization of the SHV-1 beta-Lactamase-beta-Lactamase Inhibitor Protein Interface.
J.Biol.Chem., 281, 2006
|
|
2FFY
 
 | | AmpC beta-lactamase N289A mutant in complex with a boronic acid deacylation transition state analog compound SM3 | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-PHENYLMETHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Chen, Y, Minasov, G, Roth, T.A, Prati, F, Shoichet, B.K. | | Deposit date: | 2005-12-20 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | The deacylation mechanism of AmpC beta-lactamase at ultrahigh resolution
J.Am.Chem.Soc., 128, 2006
|
|
2EFX
 
 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with L-phenylalanine amide | | Descriptor: | BARIUM ION, D-amino acid amidase, PHENYLALANINE AMIDE | | Authors: | Okazaki, S, Suzuki, A, Mizushima, T, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of D-amino-acid amidase complexed with L-phenylalanine and with L-phenylalanine amide: insight into the D-stereospecificity of D-amino-acid amidase from Ochrobactrum anthropi SV3.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2EFU
 
 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with L-phenylalanine | | Descriptor: | BARIUM ION, D-Amino acid amidase, PHENYLALANINE | | Authors: | Okazaki, S, Suzuki, A, Mizushima, T, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of D-amino-acid amidase complexed with L-phenylalanine and with L-phenylalanine amide: insight into the D-stereospecificity of D-amino-acid amidase from Ochrobactrum anthropi SV3.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2E8I
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, D1 mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Shibata, N, Higuchi, Y, Negoro, S. | | Deposit date: | 2007-01-20 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold.
Febs J., 276, 2009
|
|
2DRW
 
 | | The crystal structutre of D-amino acid amidase from Ochrobactrum anthropi SV3 | | Descriptor: | BARIUM ION, D-Amino acid amidase | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Functional Characterization of a D-Stereospecific Amino Acid Amidase from Ochrobactrum anthropi SV3, a New Member of the Penicillin-recognizing Proteins
J.Mol.Biol., 368, 2007
|
|
2DNS
 
 | | The crystal structure of D-amino acid amidase from Ochrobactrum anthropi SV3 complexed with D-Phenylalanine | | Descriptor: | BARIUM ION, D-PHENYLALANINE, D-amino acid amidase | | Authors: | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Functional Characterization of a D-Stereospecific Amino Acid Amidase from Ochrobactrum anthropi SV3, a New Member of the Penicillin-recognizing Proteins
J.Mol.Biol., 368, 2007
|
|
2DCF
 
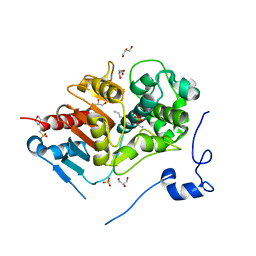 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/H266N mutant with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2006-01-06 | | Release date: | 2007-01-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Nylon-oligomer degrading enzyme/substrate complex: catalytic mechanism of 6-aminohexanoate-dimer hydrolase
J.Mol.Biol., 370, 2007
|
|
