3FFO
 
 | | F17b-G lectin domain with bound GlcNAc(beta1-2)man | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose, Adhesin, ... | | Authors: | Buts, L, De Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-12-04 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli.
Biology (Basel), 2, 2013
|
|
3F6J
 
 | | F17a-G lectin domain with bound GlcNAc(beta1-3)Gal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-methyl beta-D-galactopyranoside, F17a-G | | Authors: | Buts, L, de Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli
Biology, 2, 2013
|
|
8ARD
 
 | |
1O4S
 
 | |
2EHJ
 
 | |
2E55
 
 | |
6TXW
 
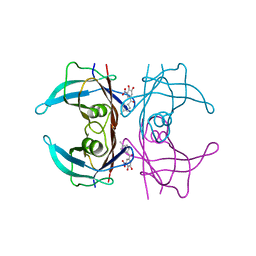 | | V30G Transthyretin structure in complex with Tolcalpone | | Descriptor: | Tolcapone, Transthyretin | | Authors: | Varejao, N, Reverter, D, Pinheiro, F, Pallares, I, Ventura, S. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.153 Å) | | Cite: | Tolcapone, a potent aggregation inhibitor for the treatment of familial leptomeningeal amyloidosis.
Febs J., 288, 2021
|
|
8AAV
 
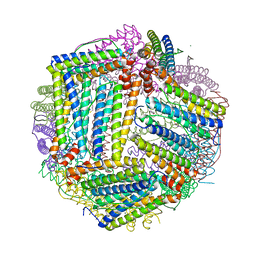 | |
3DOK
 
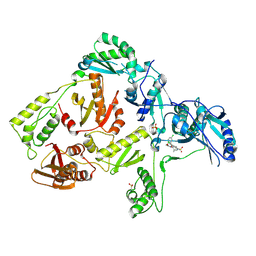 | | Crystal structure of K103N mutant HIV-1 reverse transcriptase in complex with GW678248. | | Descriptor: | 2-{4-chloro-2-[(3-chloro-5-cyanophenyl)carbonyl]phenoxy}-N-(2-methyl-4-sulfamoylphenyl)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Chamberlain, P.P, Ren, J, Stammers, D.K. | | Deposit date: | 2008-07-04 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
1GS5
 
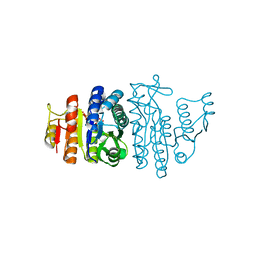 | | N-acetyl-L-glutamate kinase from Escherichia coli complexed with its substrate N-acetylglutamate and its substrate analog AMPPNP | | Descriptor: | ACETYLGLUTAMATE KINASE, MAGNESIUM ION, N-ACETYL-L-GLUTAMATE, ... | | Authors: | Ramon-Maiques, S, Marina, A, Gil-Ortiz, F, Fita, I, Rubio, V. | | Deposit date: | 2001-12-28 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Acetylglutamate Kinase, a Key Enzyme for Arginine Biosynthesis and a Prototype for the Amino Acid Kinase Enzyme Family, During Catalysis
Structure, 10, 2002
|
|
2BAL
 
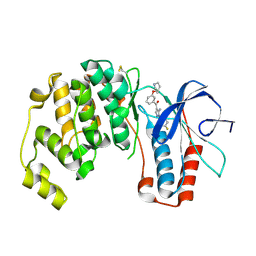 | | p38alpha MAP kinase bound to pyrazoloamine | | Descriptor: | Mitogen-activated protein kinase 14, [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL][3-(PIPERIDIN-4-YLOXY)PHENYL]METHANONE | | Authors: | Gerhardt, S, Pauptit, R.A, Read, J, Tucker, J, Norman, R.A, Breed, J. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase.
Biochemistry, 44, 2005
|
|
2BAK
 
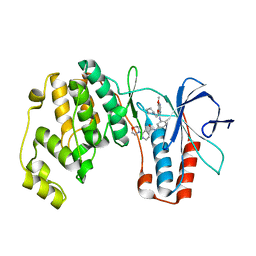 | | p38alpha MAP kinase bound to MPAQ | | Descriptor: | Mitogen-activated protein kinase 14, N-(3-{[7-METHOXY-6-(2-PYRROLIDIN-1-YLETHOXY)QUINAZOLIN-4-YL]AMINO}-4-METHYLPHENYL)-2-MORPHOLIN-4-YLISONICOTINAMIDE | | Authors: | Gerhardt, S, Breed, J, Pauptit, R.A, Read, J, Norman, R.A, Ward, W.H. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase
Biochemistry, 44, 2005
|
|
1E66
 
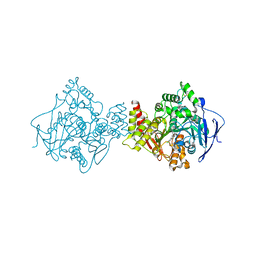 | | STRUCTURE OF ACETYLCHOLINESTERASE COMPLEXED WITH (-)-HUPRINE X AT 2.1A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-CHLORO-9-ETHYL-6,7,8,9,10,11-HEXAHYDRO-7,11-METHANOCYCLOOCTA[B]QUINOLIN-12-AMINE, ACETYLCHOLINESTERASE | | Authors: | Dvir, H, Harel, M, Silman, I, Sussman, J.L. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3D Structure of Torpedo Californica Acetylcholinesterase Complexed with Huprine X at 2. 1 A Resolution: Kinetic and Molecular Dynamic Correlates.
Biochemistry, 41, 2002
|
|
1EST
 
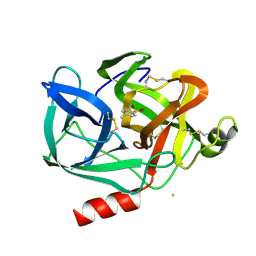 | |
2AZU
 
 | |
2BAJ
 
 | | p38alpha bound to pyrazolourea | | Descriptor: | 1-(3-tert-butyl-1-phenyl-1H-pyrazol-5-yl)-3-(2,3-dichlorophenyl)urea, Mitogen-activated protein kinase 14 | | Authors: | Gerhardt, S, Pauptit, R.A, Read, J, Breed, J, Norman, R.A, Ward, W.H. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase
Biochemistry, 44, 2005
|
|
2BAQ
 
 | | p38alpha bound to Ro3201195 | | Descriptor: | Mitogen-activated protein kinase 14, [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL](3-{[(2R)-2,3-DIHYDROXYPROPYL]OXY}PHENYL)METHANONE | | Authors: | Gerhardt, S, Pauptit, R.A, Breed, J, Read, J, Tucker, J, Norman, R.A. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Prevention of MKK6-Dependent Activation by Binding to p38alpha MAP Kinase.
Biochemistry, 44, 2005
|
|
3FYZ
 
 | | OXA-24 beta-lactamase complex with SA4-17 inhibitor | | Descriptor: | (2S,3R)-2-[(7-aminocarbonyl-2-methanoyl-indolizin-3-yl)amino]-4-aminocarbonyloxy-3-methyl-3-sulfino-butanoic acid, Beta-lactamase OXA-24, TETRAETHYLENE GLYCOL | | Authors: | Romero, A, Santillana, E. | | Deposit date: | 2009-01-23 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase.
J.Am.Chem.Soc., 132, 2010
|
|
3FZC
 
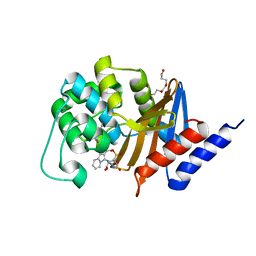 | | OXA-24 beta-lactamase complex with SA3-53 inhibitor | | Descriptor: | (2S,3R)-4-(2-aminoethylcarbamoyloxy)-2-[(2-methanoylindolizin-3-yl)amino]-3-methyl-3-sulfino-butanoic acid, Beta-lactamase OXA-24, TETRAETHYLENE GLYCOL | | Authors: | Romero, A, Santillana, E. | | Deposit date: | 2009-01-24 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase.
J.Am.Chem.Soc., 132, 2010
|
|
3FTP
 
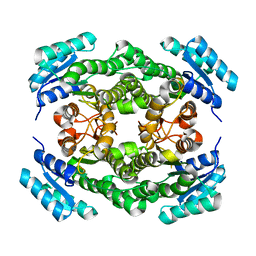 | |
3EJ2
 
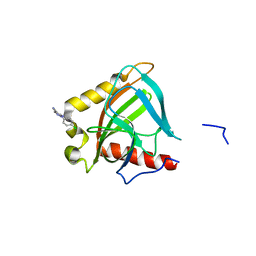 | |
1O5U
 
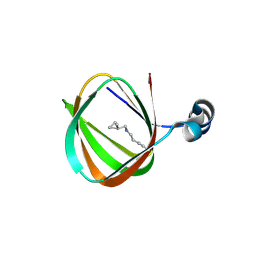 | |
3G4P
 
 | | OXA-24 beta-lactamase at pH 7.5 | | Descriptor: | Beta-lactamase OXA-24 | | Authors: | Santillana, E, Romero, A. | | Deposit date: | 2009-02-04 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase
J.Am.Chem.Soc., 132, 2010
|
|
3GGX
 
 | | HIV Protease, pseudo-symmetric inhibitors | | Descriptor: | V-1 protease, methyl [(1S)-1-{[(1R,3S,4S)-4-{[(2S)-3,3-dimethyl-2-{3-[(6-methylpyridin-2-yl)methyl]-2-oxo-2,3-dihydro-1H-imidazol-1-yl}butanoyl]amino}-3-hydroxy-5-phenyl-1-(4-pyridin-2-ylbenzyl)pentyl]carbamoyl}-2,2-dimethylpropyl]carbamate | | Authors: | Stoll, V.S. | | Deposit date: | 2009-03-02 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus
protease inhibitors (A-792611 and A-790742) with potential for convenient
dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
3GGV
 
 | | HIV Protease, pseudo-symmetric inhibitors | | Descriptor: | V-1 protease, methyl [(1S)-1-{[(1R,3S,4S)-3-hydroxy-4-{[(2S)-2-(3-{[6-(1-hydroxy-1-methylethyl)pyridin-2-yl]methyl}-2-oxo-2,3-dihydro-1H-imidazol-1-yl)-3,3-dimethylbutanoyl]amino}-5-phenyl-1-(4-pyridin-2-ylbenzyl)pentyl]carbamoyl}-2,2-dimethylpropyl]carbamate | | Authors: | Stoll, V.S. | | Deposit date: | 2009-03-02 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus
protease inhibitors (A-792611 and A-790742) with potential for convenient
dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
