2Q60
 
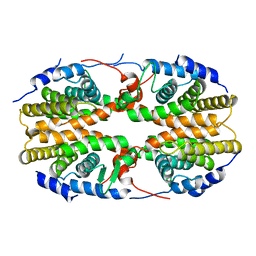 | | Crystal structure of the ligand binding domain of polyandrocarpa misakiensis rxr in tetramer in absence of ligand | | Descriptor: | Retinoid X receptor | | Authors: | Borel, F, De Groot, A, Juillan-Binard, C, De Rosny, E, Laudet, V, Pebay-Peyroula, E, Fontecilla-Camps, J.-C, Ferrer, J.-L. | | Deposit date: | 2007-06-04 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the ligand-binding domain of the retinoid X receptor from the ascidian polyandrocarpa misakiensis.
Proteins, 74, 2008
|
|
6ISA
 
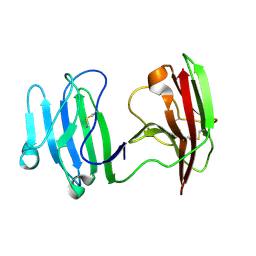 | | mCD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6UDB
 
 | | Spectroscopic and structural characterization of a genetically encoded direct sensor for protein-ligand interactions | | Descriptor: | DI(HYDROXYETHYL)ETHER, Streptavidin | | Authors: | Mills, J.H, Gleason, P.R, Simmons, C.R, Henderson, J.N, Kartchner, B.K. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Origins of Altered Spectroscopic Properties upon Ligand Binding in Proteins Containing a Fluorescent Noncanonical Amino Acid.
Biochemistry, 60, 2021
|
|
6ISB
 
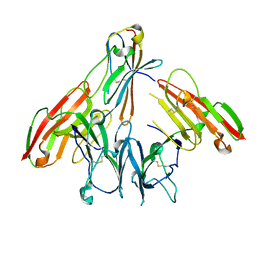 | | crystal structure of human CD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5UX6
 
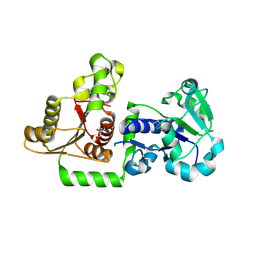 | | Structure of Human POFUT1 in its apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GLYCEROL | | Authors: | Xu, X, McMillan, B, Blacklow, S.C. | | Deposit date: | 2017-02-22 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of human POFUT1, its requirement in ligand-independent oncogenic Notch signaling, and functional effects of Dowling-Degos mutations.
Glycobiology, 27, 2017
|
|
3SE4
 
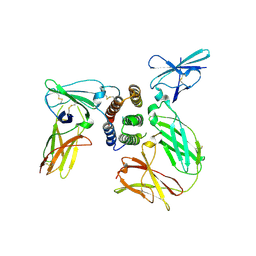 | | human IFNw-IFNAR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon alpha/beta receptor 1, Interferon alpha/beta receptor 2, ... | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5001 Å) | | Cite: | Structural linkage between ligand discrimination and receptor activation by type I interferons.
Cell(Cambridge,Mass.), 146, 2011
|
|
5YVB
 
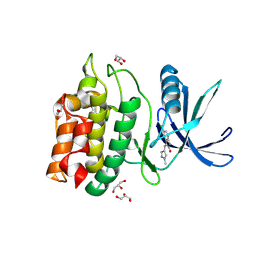 | | Structure of CaMKK2 in complex with CKI-011 | | Descriptor: | (3Z)-5-chloro-3-[(1-methyl-1H-pyrazol-4-yl)methylidene]-1,3-dihydro-2H-indol-2-one, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
1DZ9
 
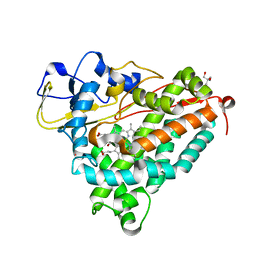 | | Putative oxo complex of P450cam from Pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
5YV9
 
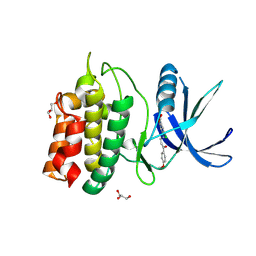 | | Structure of CaMKK2 in complex with CKI-009 | | Descriptor: | 5-chloro-2-methoxy-4[(1Z)-3-(4-methoxyphenyl)-3-oxoprop-1-en-1-yl]aminobenzoic acid, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
2J3S
 
 | |
3PGF
 
 | | Crystal structure of maltose bound MBP with a conformationally specific synthetic antigen binder (sAB) | | Descriptor: | GLYCEROL, IMIDAZOLE, Maltose-binding periplasmic protein, ... | | Authors: | Kossiakoff, A.A, Duguid, E.M, Sandstrom, A. | | Deposit date: | 2010-11-01 | | Release date: | 2011-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4B5N
 
 | | Crystal structure of oxidized Shewanella Yellow Enzyme 4 (SYE4) | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXIDOREDUCTASE, FMN-BINDING, ... | | Authors: | Elegheert, J, Brige, A, Savvides, S.N. | | Deposit date: | 2012-08-07 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural dissection of Shewanella oneidensis old yellow enzyme 4 bound to a Meisenheimer complex and (nitro)phenolic ligands.
FEBS Lett., 591, 2017
|
|
5YVA
 
 | | Structure of CaMKK2 in complex with CKI-010 | | Descriptor: | 3-(1H-tetrazol-5-yl)-10lambda~6~-thioxanthene-9,10,10-trione, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
1NAE
 
 | | Structure of CsCBM6-3 from Clostridium stercorarium in complex with xylotriose | | Descriptor: | CALCIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, putative xylanase | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilburn, D.G, Rose, D.R, Davies, G. | | Deposit date: | 2002-11-27 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and ligand binding of carbohydrate-binding module CsCBM6-3 reveals similarities with fucose-specific lectins and "galactose-binding" domains
J.Mol.Biol., 327, 2003
|
|
3S9D
 
 | | binary complex between IFNa2 and IFNAR2 | | Descriptor: | CHLORIDE ION, Interferon alpha-2, Interferon alpha/beta receptor 2 | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2011-06-01 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9999 Å) | | Cite: | Structural linkage between ligand discrimination and receptor activation by type I interferons.
Cell(Cambridge,Mass.), 146, 2011
|
|
5YV8
 
 | | Structure of CaMKK2 in complex with CKI-002 | | Descriptor: | 1-amino-4-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
4E0W
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | Kainate induces various domain closures in AMPA and kainate receptors.
Neurochem Int, 61, 2012
|
|
6C9W
 
 | | Crystal Structure of a ligand bound LacY/Nanobody Complex | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Lactose permease, Nanobody9047, ... | | Authors: | Kumar, H, Finer-Moore, J.S, Jiang, X, Smirnova, I, Kasho, V, Pardon, E, Steyaert, J, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a ligand-bound LacY-Nanobody Complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YVC
 
 | | Structure of CaMKK2 in complex with CKI-012 | | Descriptor: | 3-{2,4-dimethyl-5-[(Z)-(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-1H-pyrrol-3-yl}propanoic acid, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
8X1V
 
 | |
6DRG
 
 | | NMR solution structure of wild type hFABP1 with GW7647 | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
6A1T
 
 | | Charcot-Leyden crystal protein/Galectin-10 variant E33A with lactose | | Descriptor: | Galectin-10, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Identification of key amino acid residues determining ligand binding specificity, homodimerization and cellular distribution of human galectin-10
Glycobiology, 29, 2019
|
|
4RXA
 
 | | Crystal structure of human farnesyl diphosphate synthase in complex with BPH-1358 | | Descriptor: | Farnesyl pyrophosphate synthase, N,N'-bis[3-(4,5-dihydro-1H-imidazol-2-yl)phenyl]biphenyl-4,4'-dicarboxamide, PHOSPHATE ION | | Authors: | Liu, Y.-L, Cao, R, Wang, Y, Oldfield, E. | | Deposit date: | 2014-12-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Farnesyl diphosphate synthase inhibitors with unique ligand-binding geometries.
ACS Med Chem Lett, 6, 2015
|
|
6DQT
 
 | | Crystal structure of Haemophilus influenzae OppA complex with LGG | | Descriptor: | ACETATE ION, LEU-GLY-GLY, Periplasmic oligopeptide-binding protein, ... | | Authors: | Tanaka, K.J, Pinkett, H.W. | | Deposit date: | 2018-06-11 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Oligopeptide-binding protein from nontypeableHaemophilus influenzaehas ligand-specific sites to accommodate peptides and heme in the binding pocket.
J. Biol. Chem., 294, 2019
|
|
4RYP
 
 | |
