3RZL
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 5'-D(*AP*TP*GP*TP*AP*TP*CP*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*IP*AP*TP*AP*CP*A)-3', Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
5JFN
 
 | | Crystal structure of Rhodopseudomonas palustris propionaldehyde dehydrogenase with bound CoA and acylated Cys330 | | Descriptor: | Aldehyde dehydrogenase, COENZYME A, PENTAETHYLENE GLYCOL, ... | | Authors: | Zarzycki, J, Sutter, M, Kerfeld, C.A. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-01 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In Vitro Characterization and Concerted Function of Three Core Enzymes of a Glycyl Radical Enzyme - Associated Bacterial Microcompartment.
Sci Rep, 7, 2017
|
|
8RHE
 
 | |
8RHF
 
 | | Lytic Transglycosylase MltD of Pseudomonas aeruginosa bound to the Natural Product Bulgecin A, with two LysM domains | | Descriptor: | 4-O-(4-O-SULFONYL-N-ACETYLGLUCOSAMININYL)-5-METHYLHYDROXY-L-PROLINE-TAURINE, DI(HYDROXYETHYL)ETHER, LysM peptidoglycan-binding domain-containing protein, ... | | Authors: | Miguel-Ruano, V, Hermoso, J.A. | | Deposit date: | 2023-12-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of lytic transglycosylase MltD of Pseudomonas aeruginosa, a target for the natural product bulgecin A.
Int.J.Biol.Macromol., 267, 2024
|
|
1L3Q
 
 | | H. rufescens abalone shell Lustrin A consensus repeat, FPGKNVNCTSGE, pH 7.4, 1-H NMR structure | | Descriptor: | Lustrin A | | Authors: | Evans, J.S, Wustman, B.A, Zhang, B, Morse, D.E. | | Deposit date: | 2002-02-28 | | Release date: | 2002-03-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Model peptide studies of sequence regions in the elastomeric biomineralization protein, Lustrin A. I. The C-domain consensus-PG-, -NVNCT-motif
Biopolymers, 63, 2002
|
|
1KZX
 
 | | Solution structure of human intestinal fatty acid binding protein with a naturally-occurring single amino acid substitution (A54T) | | Descriptor: | INTESTINAL FATTY ACID-BINDING PROTEIN (T54) | | Authors: | Zhang, F, Luecke, C, Baier, L.J, Sacchettini, J.C, Hamilton, J.A. | | Deposit date: | 2002-02-08 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human intestinal fatty acid binding protein with a naturally-occurring single amino acid substitution (A54T) that is associated with altered lipid metabolism
Biochemistry, 42, 2003
|
|
9MXZ
 
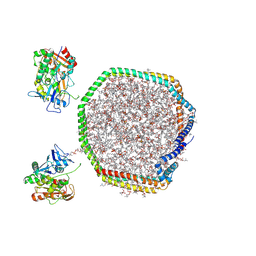 | | Lecithin:Cholesterol Acyltransferase Bound to Apolipoprotein A-I dimer in HDL | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Apolipoprotein A-I, Phosphatidylcholine-sterol acyltransferase | | Authors: | Coleman, B, Bedi, S, Hill, J.H, Morris, J, Manthei, K.A, Hart, R.C, He, Y, Shah, A.S, Jerome, W.G, Vaisar, T, Bornfeldt, K.E, Song, H, Segrest, J.P, Heinecke, J.W, Aller, S.G, Tesmer, J.J.G, Davidson, S. | | Deposit date: | 2025-01-21 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Lecithin:cholesterol acyltransferase binds a discontinuous binding site on adjacent apolipoprotein A-I belts in HDL.
J.Lipid Res., 66, 2025
|
|
3SJH
 
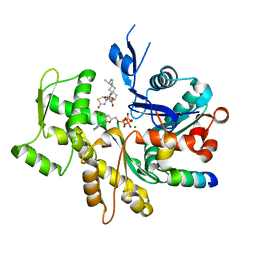 | | Crystal Structure of a chimera containing the N-terminal domain (residues 8-29) of drosophila Ciboulot and the C-terminal domain (residues 18-44) of bovine Thymosin-beta4, bound to G-actin-ATP-Latrunculin A | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-06-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly
Embo J., 31, 2012
|
|
9NM2
 
 | | Dimeric Structure of full-length CrgA, a Cell Division Protein from Mycobacterium tuberculosis, in Lipid Bilayers | | Descriptor: | Cell division protein CrgA | | Authors: | Shin, Y, Prasad, R, Das, N, Taylor, J.A, Qin, H, Hu, W, Hu, Y.-Y, Fu, R, Zhang, R, Zhou, H.-X, Cross, T.A. | | Deposit date: | 2025-03-03 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | SOLID-STATE NMR | | Cite: | Mycobacterium tuberculosis CrgA Forms a Dimeric Structure with Its Transmembrane Domain Sandwiched between Cytoplasmic and Periplasmic beta-Sheets, Enabling Multiple Interactions with Other Divisome Proteins.
J.Am.Chem.Soc., 147, 2025
|
|
5XPO
 
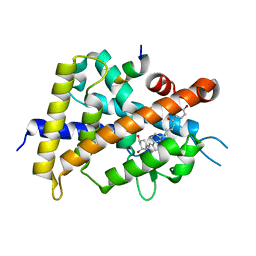 | | Crystal structure of VDR-LBD complexed with 25-(hydroxyphenyl)-2-methylidene-19,26,27-trinor-25-oxo-1-hydroxyvitamin D3 | | Descriptor: | (5~{R})-5-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl)cyclohexyl idene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]-1-(4-hydroxyphenyl)hexan-1-one, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
6EHG
 
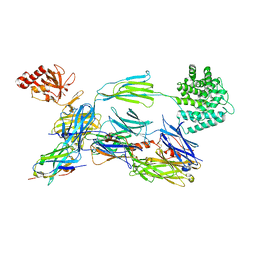 | | complement component C3b in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jensen, R.K, Andersen, K.R, Gadeberg, T.A.F, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-09-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A potent complement factor C3-specific nanobody inhibiting multiple functions in the alternative pathway of human and murine complement.
J. Biol. Chem., 293, 2018
|
|
8RHI
 
 | | Lytic Transglycosylase MltD of Pseudomonas aeruginosa in a ternary complex bound to Bulgecin A and chito-tetraose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-(4-O-SULFONYL-N-ACETYLGLUCOSAMININYL)-5-METHYLHYDROXY-L-PROLINE-TAURINE, ... | | Authors: | Miguel-Ruano, V, Hermoso, J.A. | | Deposit date: | 2023-12-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural characterization of lytic transglycosylase MltD of Pseudomonas aeruginosa, a target for the natural product bulgecin A.
Int.J.Biol.Macromol., 267, 2024
|
|
5XPP
 
 | | Crystal structure of VDR-LBD complexed with 25RS-(Hydroxyphenyl)-2-methylidene-19,26,27-trinor-1,25-dihydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{R})-6-(4-hydroxyphenyl)-6-oxidanyl-hexan-2-yl]-7~{ a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
4V9I
 
 | | Crystal structure of thermus thermophilus 70S in complex with tRNAs and mRNA containing a pseudouridine in a stop codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S Ribosomal protein S10, ... | | Authors: | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | Deposit date: | 2013-04-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
5XPM
 
 | | Crystal structure of VDR-LBD complexed with 22S-Butyl-25RS-(hydroxyphenyl)-25-methoxy-2-methylidene-19,26,27-trinor-1-hydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},3~{S})-3-[(3~{S})-3-(4-hydroxyphenyl)-3-methoxy-propyl]heptan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
7S3D
 
 | | Structure of photosystem I with bound ferredoxin from Synechococcus sp. PCC 7335 acclimated to far-red light | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2Fe-2S ferredoxin-type domain-containing protein, ... | | Authors: | Gisriel, C.J, Flesher, D.A, Shen, G, Wang, J, Ho, M, Brudvig, G.W, Bryant, D.A. | | Deposit date: | 2021-09-05 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structure of a photosystem I-ferredoxin complex from a marine cyanobacterium provides insights into far-red light photoacclimation.
J.Biol.Chem., 298, 2021
|
|
5XPL
 
 | | Crystal structure of VDR-LBD complexed with 22S-butyl-25-hydroxyphenyl-2-methylidene-19,26,27-trinor-25-oxo-1-hydroxyvitamin D3 | | Descriptor: | (4~{S})-4-[(1~{R})-1-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl )cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]ethyl]-1-(4-hydroxyphenyl)octan-1-one, Nuclear receptor coactivator 2, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
7S53
 
 | | Structure of Sortase A from Streptococcus pyogenes with the b7-b8 loop sequence from Listeria monocytogenes Sortase A | | Descriptor: | Class A sortase, sortase A chimera | | Authors: | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
7S57
 
 | | Structure of Sortase A from Streptococcus pyogenes with the b7-b8 loop sequence of Enterococcus faecalis Sortase A | | Descriptor: | Class A sortase, sortase A chimera | | Authors: | Svendsen, J.E, Johnson, D.A, Gao, M, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
1TT5
 
 | | Structure of APPBP1-UBA3-Ubc12N26: a unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8 | | Descriptor: | Ubiquitin-conjugating enzyme E2 M, ZINC ION, amyloid protein-binding protein 1, ... | | Authors: | Huang, D.T, Miller, D.W, Mathew, R, Cassell, R, Holton, J.M, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2004-06-21 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3SO3
 
 | | Structures of Fab-Protease Complexes Reveal a Highly Specific Non-Canonical Mechanism of Inhibition. | | Descriptor: | A11 FAB heavy chain, A11 FAB light chain, GLYCEROL, ... | | Authors: | Schneider, E.L, Farady, C.J, Egea, P.F, Goetz, D.H, Baharuddin, A, Craik, C.S. | | Deposit date: | 2011-06-29 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A reverse binding motif that contributes to specific protease inhibition by antibodies.
J.Mol.Biol., 415, 2012
|
|
2V8K
 
 | |
5X5I
 
 | | The X-ray crystal structure of a TetR family transcription regulator RcdA involved in the regulation of biofilm formation in Escherichia coli | | Descriptor: | HTH-type transcriptional regulator RcdA | | Authors: | Sugino, H, Usui, M, Shimada, T, Nakano, M, Ogasawara, H, Ishihama, A, Hirata, A. | | Deposit date: | 2017-02-16 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | A structural sketch of RcdA, a transcription factor controlling the master regulator of biofilm formation.
FEBS Lett., 591, 2017
|
|
1KIV
 
 | |
8RXH
 
 | | CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME WITH A/P/E-site tRNA AND mRNA : PARENTAL STRAIN | | Descriptor: | (2S)-2-[2-[4-[[(2R,3S,4S)-3-acetyloxy-4-oxidanyl-pyrrolidin-2-yl]methyl]phenoxy]ethanoylamino]-6-azanyl-hexanoic acid, 40S ribosomal protein S12, 40S ribosomal protein S14, ... | | Authors: | Rajan, K.S, Yonath, A. | | Deposit date: | 2024-02-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural and mechanistic insights into the function of Leishmania ribosome lacking a single pseudouridine modification.
Cell Rep, 43, 2024
|
|
