1JJ6
 
 | | Testing the Water-Mediated Hin Recombinase DNA Recognition by Systematic Mutations. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*CP*TP*TP*AP*TP*CP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*(5IT)P*TP*TP*TP*GP*AP*TP*AP*AP*GP*A)-3', ... | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-07-03 | | Release date: | 2002-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
1JKO
 
 | | Testing the Water-Mediated HIN Recombinase DNA Recognition by Systematic Mutations | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*CP*TP*TP*AP*CP*CP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*TP*TP*TP*TP*GP*GP*TP*AP*AP*GP*A)-3', ... | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-07-12 | | Release date: | 2002-02-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
1MAP
 
 | |
1MAQ
 
 | |
1BI5
 
 | | CHALCONE SYNTHASE FROM ALFALFA | | Descriptor: | CHALCONE SYNTHASE | | Authors: | Ferrer, J.L, Jez, J.M, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 1998-06-22 | | Release date: | 1999-06-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
7SG9
 
 | | [mC-mC] DNA mismatch in a self-assembling rhombohedral lattice at pH 5.5 | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*(5CM)P*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(5CM)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SG8
 
 | | [T-T] DNA mismatch in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Lu, B, Bernfeld, W, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7SGA
 
 | | [C-S] DNA mismatch in a self-assembling rhombohedral lattice | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*CP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(IMC)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Lu, B, Bernfeld, W, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
1FPQ
 
 | |
6B9B
 
 | |
1GED
 
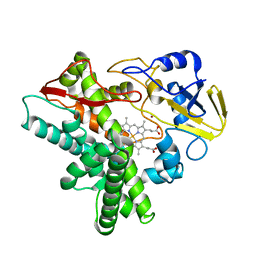 | | A positive charge route for the access of nadh to heme formed in the distal heme pocket of cytochrome p450nor | | Descriptor: | BROMIDE ION, CYTOCHROME P450 55A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kudo, T, Takaya, N, Park, S.-Y, Shiro, Y, Shoun, H. | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A positively charged cluster formed in the heme-distal pocket of cytochrome P450nor is essential for interaction with NADH
J.Biol.Chem., 276, 2001
|
|
2VK6
 
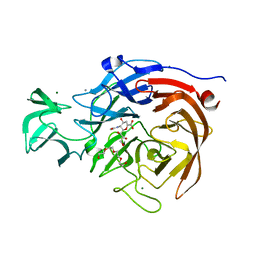 | | THE STRUCTURE OF CLOSTRIDIUM PERFRINGENS NANI SIALIDASE AND ITS CATALYTIC INTERMEDIATES | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, EXO-ALPHA-SIALIDASE, ... | | Authors: | Newstead, S.L, Potter, J.A, Wilson, J.C, Xu, G, Chien, C.H, Watts, A.G, Withers, S.G, Taylor, G.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of Clostridium Perfringens Nani Sialidase and its Catalytic Intermediates.
J.Biol.Chem., 283, 2008
|
|
5JYC
 
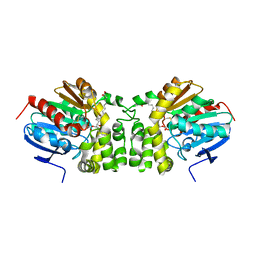 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 14,15-EET hydrolysis intermediate | | Descriptor: | (5~{Z},11~{Z},14~{R},15~{R})-14,15-bis(oxidanyl)icosa-5,8,11-trienoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-05-13 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pseudomonas aeruginosa sabotages the generation of host proresolving lipid mediators.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7KCX
 
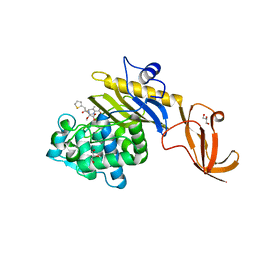 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) mutant (R200L) in complex with cefoxitin | | Descriptor: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2020-10-07 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
7KCW
 
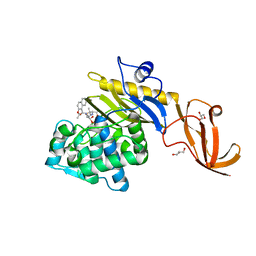 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) mutant (R200L) in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2-ethoxynaphthalen-1-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | Authors: | Alexander, J.A, Strynadka, N.C. | | Deposit date: | 2020-10-07 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
7KCY
 
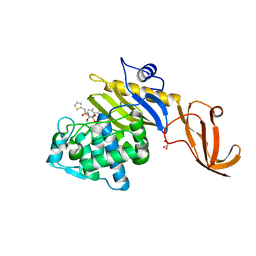 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) with cefoxitin | | Descriptor: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2020-10-07 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
1CHW
 
 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH HEXANOYL-COA | | Descriptor: | HEXANOYL-COENZYME A, PROTEIN (CHALCONE SYNTHASE) | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-30 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
2VLF
 
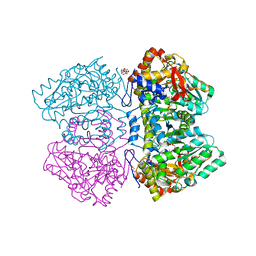 | | Quinonoid intermediate of Citrobacter freundii tyrosine phenol-lyase formed with alanine | | Descriptor: | (2E)-2-{[(Z)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4(1H)-YLIDENE}METHYL]IMINO}PROPANOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Milic, D, Demidkina, T.V, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2008-01-14 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Insights Into the Catalytic Mechanism of Tyrosine Phenol-Lyase from X-Ray Structures of Quinonoid Intermediates.
J.Biol.Chem., 283, 2008
|
|
1GPU
 
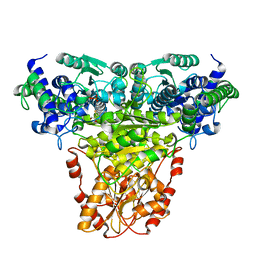 | | Transketolase complex with reaction intermediate | | Descriptor: | 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, TRANSKETOLASE 1 | | Authors: | Fiedler, E, Thorell, S, Sandalova, T, Koenig, S, Schneider, G. | | Deposit date: | 2001-11-09 | | Release date: | 2002-02-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Snapshot of a Key Intermediate in Enzymatic Thiamin Catalysis: Crystal Structure of the Alpha-Carbanion of (Alpha,Beta-Dihydroxyethyl)-Thiamin Diphosphate in the Active Site of Transketolase from Saccharomyces Cerevisiae.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1CML
 
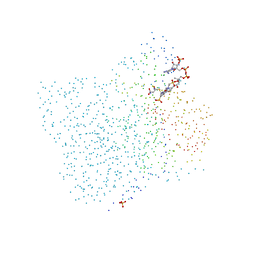 | | CHALCONE SYNTHASE FROM ALFALFA COMPLEXED WITH MALONYL-COA | | Descriptor: | MALONYL-COENZYME A, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), PROTEIN (CHALCONE SYNTHASE), ... | | Authors: | Ferrer, J.-L, Jez, J, Bowman, M.E, Dixon, R, Noel, J.P. | | Deposit date: | 1999-03-30 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
4NYM
 
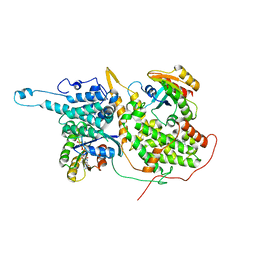 | | Approach for Targeting Ras with Small Molecules that Activate SOS-Mediated Nucleotide Exchange | | Descriptor: | GTPase HRas, MAGNESIUM ION, N-[1-(1H-indol-3-ylmethyl)piperidin-4-yl]-L-tryptophanamide, ... | | Authors: | Burns, M.C, Sun, Q, Phan, J, Fesik, S.W. | | Deposit date: | 2013-12-10 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5529 Å) | | Cite: | Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3C5E
 
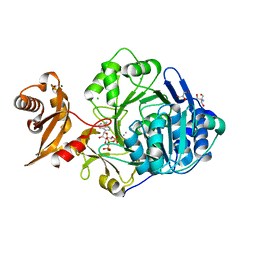 | | Crystal structure of human acyl-CoA synthetase medium-chain family member 2A (L64P mutation) in complex with ATP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, Acyl-coenzyme A synthetase ACSM2A, ... | | Authors: | Pilka, E.S, Kochan, G.T, Bhatia, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural snapshots for the conformation-dependent catalysis by human medium-chain acyl-coenzyme A synthetase ACSM2A.
J.Mol.Biol., 388, 2009
|
|
1BQ6
 
 | | CHALCONE SYNTHASE FROM ALFALFA WITH COENZYME A | | Descriptor: | CHALCONE SYNTHASE, COENZYME A, SULFATE ION | | Authors: | Ferrer, J.-L, Bowman, M.E, Jez, J, Dixon, R, Noel, J.P. | | Deposit date: | 1998-08-21 | | Release date: | 1999-08-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
4NYJ
 
 | | Approach for Targeting Ras with Small Molecules that Activate SOS-Mediated Nucleotide Exchange | | Descriptor: | GTPase HRas, MAGNESIUM ION, N-[1-(1H-indol-3-ylmethyl)piperidin-4-yl]glycinamide, ... | | Authors: | Burns, M.C, Sun, Q, Phan, J, Fesik, S.W. | | Deposit date: | 2013-12-10 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8522 Å) | | Cite: | Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NYI
 
 | | Approach for Targeting Ras with Small Molecules that Activate SOS-Mediated Nucleotide Exchange | | Descriptor: | GTPase HRas, MAGNESIUM ION, N-{1-[(5-methyl-1H-indol-3-yl)methyl]piperidin-4-yl}-L-tryptophanamide, ... | | Authors: | Burns, M.C, Sun, Q, Phan, J, Fesik, S.W. | | Deposit date: | 2013-12-10 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9612 Å) | | Cite: | Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
