2QM4
 
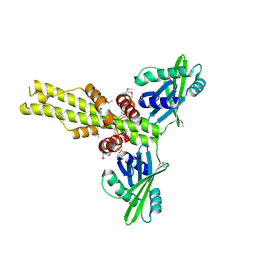 | | Crystal structure of human XLF/Cernunnos, a non-homologous end-joining factor | | Descriptor: | Non-homologous end-joining factor 1 | | Authors: | Li, Y, Chirgadze, D.Y, Sibanda, B.L, Bolanos-Garcia, V.M, Davies, O.R, Blundell, T.L. | | Deposit date: | 2007-07-14 | | Release date: | 2007-12-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ.
Embo J., 27, 2008
|
|
1DRG
 
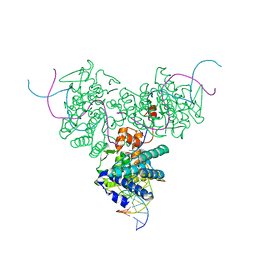 | | CRYSTAL STRUCTURE OF TRIMERIC CRE RECOMBINASE-LOX COMPLEX | | Descriptor: | 5'-D(*AP*TP*AP*TP*GP*CP*TP*AP*TP*AP*CP*GP*AP*AP*GP*TP*TP*AP*T)-3', 5'-D(*TP*AP*TP*AP*AP*CP*TP*TP*CP*GP*TP*AP*TP*AP*GP*C)-3', CRE RECOMBINASE | | Authors: | Woods, K.C, Baldwin, E.P. | | Deposit date: | 2000-01-06 | | Release date: | 2001-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Quasi-equivalence in site-specific recombinase structure and function: crystal structure and activity of trimeric Cre recombinase bound to a three-way Lox DNA junction
J.Mol.Biol., 313, 2001
|
|
6CIL
 
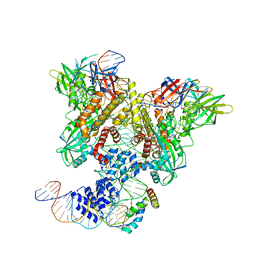 | | PRE-REACTION COMPLEX, RAG1(E962Q)/2-INTACT/INTACT 12/23RSS COMPLEX IN MN2+ | | Descriptor: | High mobility group protein B1, Intact 12RSS substrate forward strand, Intact 12RSS substrate reverse strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
8PBC
 
 | |
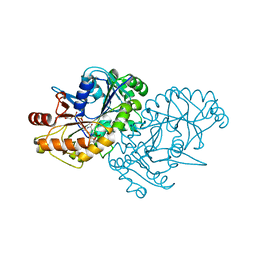
7ODX
 
 | |
8PBD
 
 | |
2A4D
 
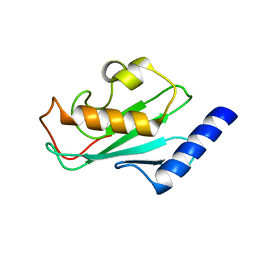 | | Structure of the human ubiquitin-conjugating enzyme E2 variant 1 (UEV-1) | | Descriptor: | Ubiquitin-conjugating enzyme E2 variant 1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-28 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
7P3A
 
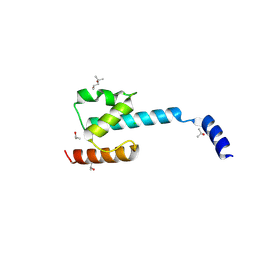 | | N-terminal domain of CGI-99 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Kroupova, A, Jinek, M. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular architecture of the human tRNA ligase complex.
Elife, 10, 2021
|
|
2PI2
 
 | | Full-length Replication protein A subunits RPA14 and RPA32 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit | | Authors: | Deng, X, Borgstahl, G.E. | | Deposit date: | 2007-04-12 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Full-length Human RPA14/32 Complex Gives Insights into the Mechanism of DNA Binding and Complex Formation.
J.Mol.Biol., 374, 2007
|
|
2BAY
 
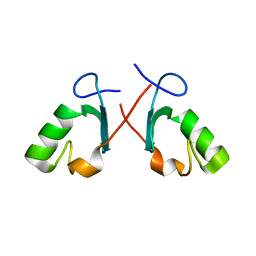 | | Crystal structure of the Prp19 U-box dimer | | Descriptor: | Pre-mRNA splicing factor PRP19 | | Authors: | Vander Kooi, C.W, Ohi, M.D, Rosenberg, J.A, Oldham, M.L, Newcomer, M.E, Gould, K.L, Chazin, W.J. | | Deposit date: | 2005-10-15 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Prp19 U-box Crystal Structure Suggests a Common Dimeric Architecture for a Class of Oligomeric E3 Ubiquitin Ligases.
Biochemistry, 45, 2006
|
|
5M8C
 
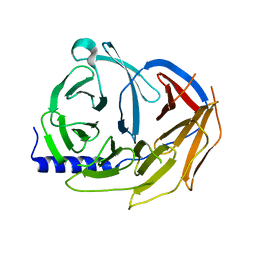 | | Spliceosome component | | Descriptor: | Pre-mRNA-processing factor 19 | | Authors: | Moura, T.R, Pena, V. | | Deposit date: | 2016-10-28 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
Mol. Cell, 69, 2018
|
|
5M88
 
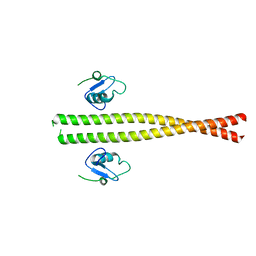 | | Spliceosome component | | Descriptor: | Core | | Authors: | Moura, T.R, Pena, V. | | Deposit date: | 2016-10-28 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
Mol. Cell, 69, 2018
|
|
5M89
 
 | | Spliceosome component | | Descriptor: | Spliceosome WD40 Sc | | Authors: | Moura, T.R, Pena, V. | | Deposit date: | 2016-10-28 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
Mol. Cell, 69, 2018
|
|
3M62
 
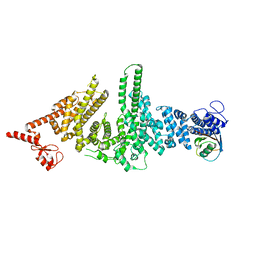 | |
5V1A
 
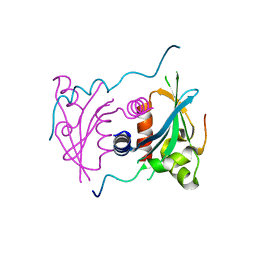 | | Structure of S. cerevisiae Ulp2:Csm1 complex | | Descriptor: | Monopolin complex subunit CSM1, Ubiquitin-like-specific protease 2 | | Authors: | Singh, N, Corbett, K.D. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Recruitment of a SUMO isopeptidase to rDNA stabilizes silencing complexes by opposing SUMO targeted ubiquitin ligase activity.
Genes Dev., 31, 2017
|
|
5V3N
 
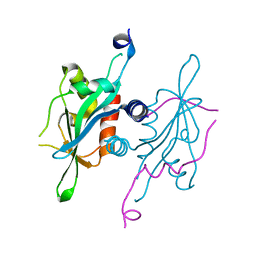 | | Structure of S. cerevisiae Ulp2-Tof2-Csm1 complex | | Descriptor: | Monopolin complex subunit CSM1, Ulp2p,Topoisomerase 1-associated factor 2 chimera | | Authors: | SIngh, N, Corbett, K.D. | | Deposit date: | 2017-03-07 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Recruitment of a SUMO isopeptidase to rDNA stabilizes silencing complexes by opposing SUMO targeted ubiquitin ligase activity.
Genes Dev., 31, 2017
|
|
7BGB
 
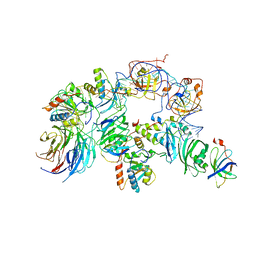 | | The H/ACA RNP lobe of human telomerase | | Descriptor: | H/ACA ribonucleoprotein complex subunit 1, H/ACA ribonucleoprotein complex subunit 2, H/ACA ribonucleoprotein complex subunit 3, ... | | Authors: | Nguyen, T.H.D, Ghanim, G.E, Fountain, A.J, van Roon, A.M.M, Rangan, R, Das, R, Collins, K. | | Deposit date: | 2021-01-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human telomerase holoenzyme with bound telomeric DNA.
Nature, 593, 2021
|
|
5YY9
 
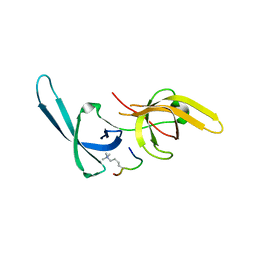 | | Crystal structure of Tandem Tudor Domain of human UHRF1 in complex with LIG1-K126me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Ligase 1 | | Authors: | Kori, S, Defossez, P.A, Arita, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
2YB6
 
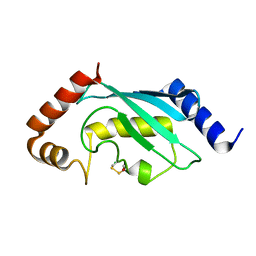 | | Native human Rad6 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, UBIQUITIN-CONJUGATING ENZYME E2 B | | Authors: | Hibbert, R.G, Sixma, T.K. | | Deposit date: | 2011-03-02 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | E3 Ligase Rad18 Promotes Monoubiquitination Rather Than Ubiquitin Chain Formation by E2 Enzyme Rad6.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8QE8
 
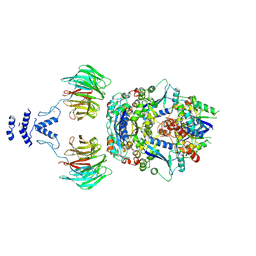 | | Structure of the non-canonical CTLH E3 substrate receptor WDR26 bound to NMNAT1 substrate | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide/nicotinic acid mononucleotide adenylyltransferase 1, WD repeat-containing protein 26, ... | | Authors: | Chrustowicz, J, Sherpa, D, Schulman, B.A. | | Deposit date: | 2023-08-30 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Non-canonical substrate recognition by the human WDR26-CTLH E3 ligase regulates prodrug metabolism.
Mol.Cell, 84, 2024
|
|
4Y18
 
 | |
4Y2G
 
 | |
6K08
 
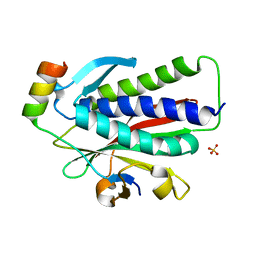 | | Crystal structure of REV7(R124A/A135D) in complex with a Shieldin3 fragment | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, SULFATE ION, Shieldin complex subunit 3 | | Authors: | Zhang, F, Dai, Y. | | Deposit date: | 2019-05-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structural basis for shieldin complex subunit 3-mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair.
J.Biol.Chem., 295, 2020
|
|
6K07
 
 | | Crystal structure of REV7(R124A) in complex with a Shieldin3 fragment | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, SULFATE ION, Shieldin complex subunit 3 | | Authors: | Zhang, F, Dai, Y. | | Deposit date: | 2019-05-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for shieldin complex subunit 3-mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair.
J.Biol.Chem., 295, 2020
|
|
8A58
 
 | |
