5WRO
 
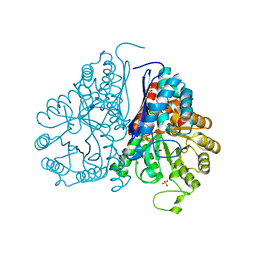 | | Crystal structure of Drosophila enolase | | Descriptor: | CADMIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Zhang, Z, Shi, Z. | | Deposit date: | 2016-12-02 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Crystal structure of enolase from Drosophila melanogaster.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
7Y47
 
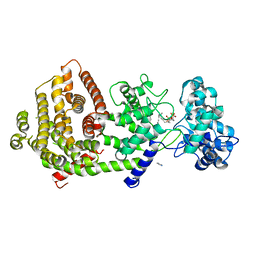 | | Crystal structure of bifunctional miltiradiene synthase from selaginella moellendorffii that complexed with GGPP | | Descriptor: | Bifunctional diterpene synthase, chloroplastic, GERANYLGERANYL DIPHOSPHATE, ... | | Authors: | Ma, X, Tao, Y, Jiang, T. | | Deposit date: | 2022-06-14 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mechanistic insights into the precise product synthesis by a bifunctional miltiradiene synthase.
Plant Biotechnol J, 21, 2023
|
|
7WAT
 
 | |
6I0Y
 
 | | TnaC-stalled ribosome complex with the titin I27 domain folding close to the ribosomal exit tunnel | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Su, T, Kudva, R, von Heijne, G, Beckmann, R. | | Deposit date: | 2018-10-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Folding pathway of an Ig domain is conserved on and off the ribosome.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6WEO
 
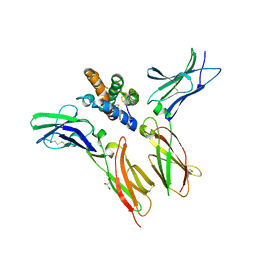 | | IL-22 Signaling Complex with IL-22R1 and IL-10Rbeta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saxton, R.A, Jude, K.M, Henneberg, L.T, Garcia, K.C. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The tissue protective functions of interleukin-22 can be decoupled from pro-inflammatory actions through structure-based design.
Immunity, 54, 2021
|
|
5HW0
 
 | |
2YBA
 
 | | Crystal structure of Nurf55 in complex with histone H3 | | Descriptor: | HISTONE H3, PROBABLE HISTONE-BINDING PROTEIN CAF1 | | Authors: | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks
Mol.Cell, 42, 2011
|
|
6ASZ
 
 | |
6AT0
 
 | |
6E3K
 
 | | Interferon gamma signalling complex with IFNGR1 and IFNGR2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jude, K.M, Mendoza, J.L, Garcia, K.C. | | Deposit date: | 2018-07-14 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the IFN gamma receptor complex guides design of biased agonists.
Nature, 567, 2019
|
|
6N9E
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with a short substrate mimic CC-Pmn and bound to mRNA and P-site tRNA at 3.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S19, ... | | Authors: | Melnikov, S.V, Khabibullina, N.F, Mairhofer, E, Vargas-Rodriguez, O, Reynolds, N.M, Micura, R, Soll, D, Polikanov, Y.S. | | Deposit date: | 2018-12-03 | | Release date: | 2018-12-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanistic insights into the slow peptide bond formation with D-amino acids in the ribosomal active site.
Nucleic Acids Res., 47, 2019
|
|
6N9F
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with a short substrate mimic ACCA-DPhe and bound to mRNA and P-site tRNA at 3.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S19, ... | | Authors: | Melnikov, S.V, Khabibullina, N.F, Mairhofer, E, Vargas-Rodriguez, O, Reynolds, N.M, Micura, R, Soll, D, Polikanov, Y.S. | | Deposit date: | 2018-12-03 | | Release date: | 2018-12-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanistic insights into the slow peptide bond formation with D-amino acids in the ribosomal active site.
Nucleic Acids Res., 47, 2019
|
|
6X7F
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B2 (TTC-B2) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6X9Q
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 27 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6X7K
 
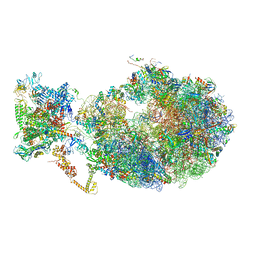 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-05-30 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XDQ
 
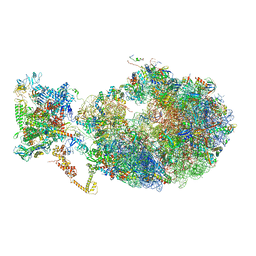 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 30 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-06-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
5JJ0
 
 | | Structure of G9a SET-domain with Histone H3K9M peptide and excess SAH | | Descriptor: | Histone H3K9M mutant peptide, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JIY
 
 | | Structure of G9a SET-domain with Histone H3K9norLeucine mutant peptide and bound S-adenosylmethionine | | Descriptor: | Histone H3.1 mutant peptide with H3K9nor-leucine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JHN
 
 | | Structure of G9a SET-domain with Histone H3K9Ala mutant peptide and bound S-adenosylmethionine | | Descriptor: | Histone H3.1 peptide with K9A mutation, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-21 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JIN
 
 | | Structure of G9a SET-domain with Histone H3K9M mutant peptide and bound S-adenosylmethionine | | Descriptor: | Histone H3.1 peptide with K9M mutation, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3P7J
 
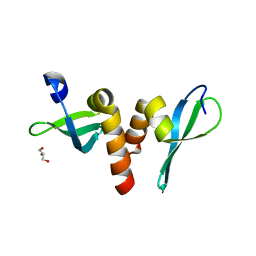 | | Drosophila HP1a chromo shadow domain | | Descriptor: | GLYCEROL, Heterochromatin protein 1 | | Authors: | Kim, D, Chruszcz, M, Minor, W, Khorasanizadeh, S. | | Deposit date: | 2010-10-12 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The HP1a Disordered C Terminus and Chromo Shadow Domain Cooperate to Select Target Peptide Partners.
Chembiochem, 12, 2011
|
|
1Q3L
 
 | |
9EV2
 
 | |
7P3K
 
 | | Cryo-EM structure of 70S ribosome stalled with TnaC peptide (control) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Buschauer, R, Komar, T, Becker, T, Berninghausen, O, Cheng, J, Beckmann, R. | | Deposit date: | 2021-07-08 | | Release date: | 2021-10-27 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of l-tryptophan-dependent inhibition of release factor 2 by the TnaC arrest peptide.
Nucleic Acids Res., 49, 2021
|
|
1MVX
 
 | | structure of the SET domain histone lysine methyltransferase Clr4 | | Descriptor: | CRYPTIC LOCI REGULATOR 4, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Min, J.R, Zhang, X, Cheng, X.D, Grewal, S.I.S, Xu, R.-M. | | Deposit date: | 2002-09-26 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the SET domain histone lysine methyltransferase Clr4.
Nat.Struct.Biol., 9, 2002
|
|
