6FDU
 
 | | Structure of Chlamydia trachomatis effector protein Cdu1 bound to Compound 3 | | Descriptor: | (2~{S},3~{S})-2-[[(2~{S})-2-[3,5-bis(chloranyl)phenyl]-2-(dimethylamino)ethanoyl]amino]-~{N}-[[2-(iminomethyl)pyrimidin-4-yl]methyl]-3-methyl-pentanamide, CHLORIDE ION, Deubiquitinase and deneddylase Dub1 | | Authors: | Ramirez, Y, Kisker, C, Altmann, E. | | Deposit date: | 2017-12-26 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Substrate Recognition and Covalent Inhibition of Cdu1 from Chlamydia trachomatis.
ChemMedChem, 13, 2018
|
|
6FDQ
 
 | |
4RIS
 
 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | Descriptor: | CH58-UA Fab heavy chain, CH58-UA Fab light chain, Envelope glycoprotein | | Authors: | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
4RIR
 
 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | Descriptor: | CH58-UA Fab heavy chain, CH58-UA Fab light chain | | Authors: | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
7E4U
 
 | |
4NEW
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma cruzi in complex with inhibitor EP127 (5-{5-[1-(PYRROLIDIN-1-YL)CYCLOHEXYL]-1,3-THIAZOL-2-YL}-1H-INDOLE) | | Descriptor: | 5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, Trypanothione reductase, ... | | Authors: | Persch, E, Bryson, S, Pai, E.F, Krauth-Siegel, R.L, Diederich, F. | | Deposit date: | 2013-10-30 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding to large enzyme pockets: small-molecule inhibitors of trypanothione reductase.
Chemmedchem, 9, 2014
|
|
4NEV
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor EP127 (5-{5-[1-(PYRROLIDIN-1-YL)CYCLOHEXYL]-1,3-THIAZOL-2-YL}-1H-INDOLE) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Persch, E, Bryson, S, Pai, E.F, Krauth-Siegel, R.L, Diederich, F. | | Deposit date: | 2013-10-30 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding to large enzyme pockets: small-molecule inhibitors of trypanothione reductase.
Chemmedchem, 9, 2014
|
|
2UYL
 
 | | Crystal structure of a monoclonal antibody directed against an antigenic determinant common to Ogawa and Inaba serotypes of Vibrio cholerae O1 | | Descriptor: | MONOCLONAL ANTIBODY F-22-30 | | Authors: | Ahmed, F, Haouz, A, Nato, F, Fournier, J.M, Alzari, P.M. | | Deposit date: | 2007-04-10 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Monoclonal Antibody Directed Against an Antigenic Determinant Common to Ogawa and Inaba Serotypes of Vibrio Cholerae O1.
Proteins, 70, 2008
|
|
2V2V
 
 | | IspE in complex with ligand | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2C-METHYL-D-ERYTHRITOL KINASE, 5'-[(1H-BENZIMIDAZOL-2-YLACETYL)AMINO]-5'-DEOXYCYTIDINE, BROMIDE ION, ... | | Authors: | Alphey, M.S, Hunter, W.N. | | Deposit date: | 2007-06-07 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and Characterization of Cytidine Derivatives that Inhibit the Kinase Ispe of the Non-Mevalonate Pathway for Isoprenoid Biosynthesis.
Chemmedchem, 3, 2008
|
|
2V2Q
 
 | | IspE in complex with ligand | | Descriptor: | 4-AMINO-1-(5-{[3-(1H-BENZIMIDAZOL-2-YL)PROPANOYL]AMINO}-5-DEOXY-ALPHA-L-LYXOFURANOSYL)PYRIMIDIN-2(1H)-ONE, 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL KINASE, BROMIDE ION, ... | | Authors: | Alphey, M.S, Hunter, W.N. | | Deposit date: | 2007-06-06 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and Characterization of Cytidine Derivatives that Inhibit the Kinase Ispe of the Non-Mevalonate Pathway for Isoprenoid Biosynthesis.
Chemmedchem, 3, 2008
|
|
2GGF
 
 | | Solution structure of the MA3 domain of human Programmed cell death 4 | | Descriptor: | Programmed cell death 4, isoform 1 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-24 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the MA3 domain of human Programmed cell death 4
To be Published
|
|
2JWQ
 
 | | G-quadruplex recognition by quinacridines: a SAR, NMR and Biological study | | Descriptor: | DNA (5'-D(*DTP*DTP*DAP*DGP*DGP*DGP*DT)-3'), N,N'-(dibenzo[b,j][1,7]phenanthroline-2,10-diyldimethanediyl)dipropan-1-amine | | Authors: | Hounsou, C, Guittat, L, Monchaud, D, Jourdan, M, Saettel, N, Mergny, J.L, Teulade-Fichou, M. | | Deposit date: | 2007-10-23 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | G-Quadruplex Recognition by Quinacridines: a SAR, NMR, and Biological Study
ChemMedChem, 2, 2007
|
|
3GCM
 
 | |
2NUH
 
 | |
4INB
 
 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With benzodiazepine Inhibitor | | Descriptor: | (3Z)-3-{[(2-methoxyethyl)amino]methylidene}-1-methyl-5-phenyl-7-(trifluoromethyl)-1H-1,5-benzodiazepine-2,4(3H,5H)-dione, Gag protein, SODIUM ION | | Authors: | Coulombe, R. | | Deposit date: | 2013-01-04 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Monitoring Binding of HIV-1 Capsid Assembly Inhibitors Using (19) F Ligand-and (15) N Protein-Based NMR and X-ray Crystallography: Early Hit Validation of a Benzodiazepine Series.
Chemmedchem, 8, 2013
|
|
3GKQ
 
 | |
2HRC
 
 | | 1.7 angstrom structure of human ferrochelatase variant R115L | | Descriptor: | CHLORIDE ION, CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Medlock, A, Swartz, L, Dailey, T.A, Dailey, H.A, Lanzilotta, W.N. | | Deposit date: | 2006-07-20 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate interactions with human ferrochelatase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3HCO
 
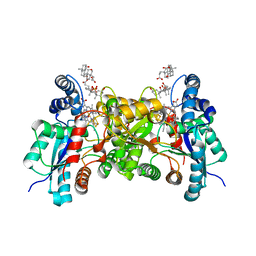 | | Human ferrochelatase with Cd and protoporphyrin IX bound | | Descriptor: | BICARBONATE ION, CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Medlock, A.E, Dailey, H.A, Lanzilotta, W.N. | | Deposit date: | 2009-05-06 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Product release rather than chelation determines metal specificity for ferrochelatase.
J.Mol.Biol., 393, 2009
|
|
3GME
 
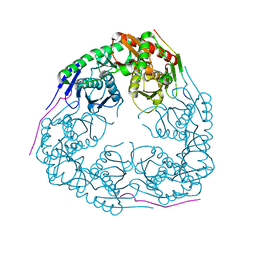 | |
3GLL
 
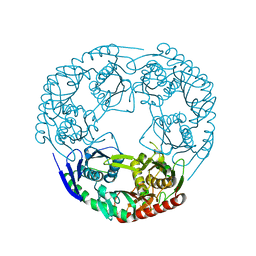 | | Crystal structure of Polynucleotide Phosphorylase (PNPase) core | | Descriptor: | Polyribonucleotide nucleotidyltransferase | | Authors: | Nurmohamed, S, Luisi, B.L. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Escherichia coli polynucleotide phosphorylase core bound to RNase E, RNA and manganese: implications for catalytic mechanism and RNA degradosome assembly.
J.Mol.Biol., 389, 2009
|
|
3HCR
 
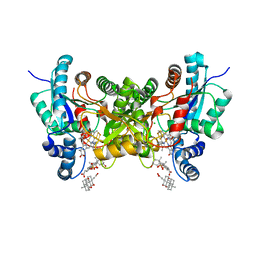 | | Human Ferrochelatase with deuteroporphyrin and Ni Bound | | Descriptor: | CHLORIDE ION, CHOLIC ACID, FE(III) DEUTEROPORPHYRIN IX, ... | | Authors: | Medlock, A.E, Dailey, H.A, Lanzilotta, W.N. | | Deposit date: | 2009-05-06 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Product release rather than chelation determines metal specificity for ferrochelatase.
J.Mol.Biol., 393, 2009
|
|
3HCN
 
 | | Hg and protoporphyrin bound Human Ferrochelatase | | Descriptor: | BICARBONATE ION, CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Medlock, A.E, Dailey, H.A, Lanzilotta, W.N. | | Deposit date: | 2009-05-06 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Product release rather than chelation determines metal specificity for ferrochelatase.
J.Mol.Biol., 393, 2009
|
|
3HCP
 
 | | Human ferrochelatase with Mn and deuteroporphyrin bound | | Descriptor: | CHOLIC ACID, FE(III) DEUTEROPORPHYRIN IX, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Medlock, A.E, Dailey, H.A, Lanzilotta, W.N. | | Deposit date: | 2009-05-06 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Product release rather than chelation determines metal specificity for ferrochelatase.
J.Mol.Biol., 393, 2009
|
|
7MBO
 
 | | FACTOR XIA (PICHIA PASTORIS; C500S [C122S]) IN COMPLEX WITH THE INHIBITOR Milvexian (BMS-986177), IUPAC NAME:(6R,10S)-10-{4-[5-chloro-2-(4-chloro-1H-1,2,3-triazol-1-yl)phenyl]-6- oxopyrimidin-1(6H)-yl}-1-(difluoromethyl)-6-methyl-1,4,7,8,9,10-hexahydro-15,11- (metheno)pyrazolo[4,3-b][1,7]diazacyclotetradecin-5(6H)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, Milvexian | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.924 Å) | | Cite: | Discovery of Milvexian, a High-Affinity, Orally Bioavailable Inhibitor of Factor XIa in Clinical Studies for Antithrombotic Therapy.
J.Med.Chem., 65, 2022
|
|
4U4S
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM25 at 1.90 A resolution. | | Descriptor: | 4-ethyl-3,4-dihydro-2H-pyrido[4,3-e][1,2,4]thiadiazine 1,1-dioxide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Noerholm, A.B, Deva, T, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2014-07-24 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Positive Allosteric Modulators of 2-Amino-3-(3-hydroxy-5-methylisoxazol-4-yl)propionic Acid Receptors Belonging to 4-Cyclopropyl-3,4-dihydro-2H-1,2,4-pyridothiadiazine Dioxides and Diversely Chloro-Substituted 4-Cyclopropyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides.
J.Med.Chem., 57, 2014
|
|
