4Y32
 
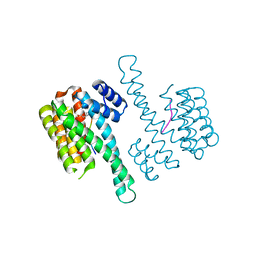 | | Crystal structure of C-terminal modified Tau peptide-hybrid 109B with 14-3-3sigma | | Descriptor: | (2S)-2-(2-methoxyethyl)pyrrolidine, 14-3-3 protein sigma, ARG-THR-PRO-SEP-LEU-PRO-CNC(C(C)O)C(=O)N1CCCC1CCOC | | Authors: | Bartel, M, Milroy, L, Bier, D, Brunsveld, L, Ottmann, C. | | Deposit date: | 2015-02-10 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Stabilizer-Guided Inhibition of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XRS
 
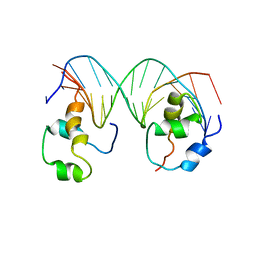 | | Heterodimeric complex of transcription factors MEIS1 and DLX3 on specific DNA | | Descriptor: | DNA (5'-D(P*AP*CP*AP*AP*TP*TP*AP*TP*CP*CP*TP*GP*TP*CP*AP*AP*C)-3'), DNA (5'-D(P*CP*AP*AP*TP*TP*AP*TP*CP*CP*TP*GP*TP*CP*AP*A)-3'), DNA (5'-D(P*GP*TP*TP*GP*AP*CP*AP*GP*GP*AP*TP*AP*AP*TP*TP*GP*TP*T)-3'), ... | | Authors: | Jorma, A, Yin, Y, Nitta, K.R, Dave, K, Enge, M, Kivioja, T, Popov, A, Morgunova, E, Taipale, J. | | Deposit date: | 2015-01-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | DNA-dependent formation of transcription factor pairs alters their binding specificity.
Nature, 527, 2015
|
|
4XZV
 
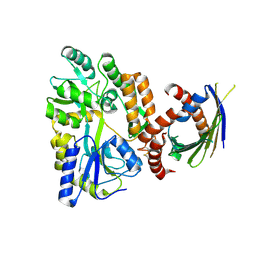 | | Crystal Structure of SLMO1-TRIAP1 Complex | | Descriptor: | Maltose-binding periplasmic protein,TP53-regulated inhibitor of apoptosis 1, Protein slowmo homolog 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Miliara, X, Garnett, J.A, Matthews, S.J. | | Deposit date: | 2015-02-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes.
Embo Rep., 16, 2015
|
|
1TX4
 
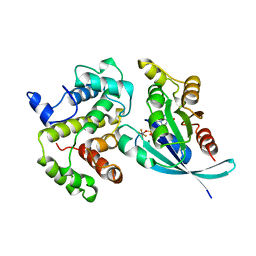 | | RHO/RHOGAP/GDP(DOT)ALF4 COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, P50-RHOGAP, ... | | Authors: | Rittinger, K, Walker, P.A, Smerdon, S.J, Gamblin, S.J. | | Deposit date: | 1997-07-29 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue.
Nature, 389, 1997
|
|
3KBZ
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 6 | | Descriptor: | Fructose-1,6-bisphosphatase 1, {[(2-amino-8H-indeno[1,2-d][1,3]thiazol-4-yl)oxy]methyl}phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5MPH
 
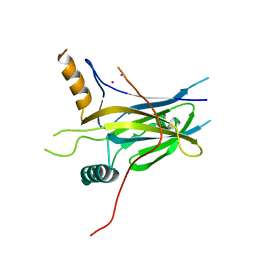 | | Structural Basis of Gene Regulation by the Grainyhead Transcription Factor Superfamily | | Descriptor: | Grainyhead-like protein 1 homolog, IODIDE ION | | Authors: | Ming, Q, Roske, Y, Schuetz, A, Walentin, K, Ibraimi, I, Schmidt-Ott, K.M, Heinemann, U. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.337 Å) | | Cite: | Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family.
Nucleic Acids Res., 46, 2018
|
|
3KC1
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 19a | | Descriptor: | Fructose-1,6-bisphosphatase 1, {[(7-carbamoyl-8H-indeno[1,2-d][1,3]thiazol-4-yl)oxy]methyl}phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KD0
 
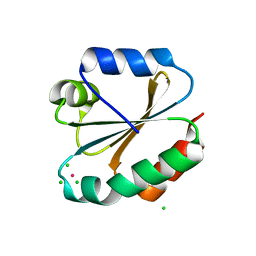 | |
5MYC
 
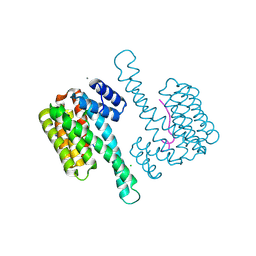 | | Crystal structure of human 14-3-3 sigma in complex with LRRK2 peptide pS910 | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stevers, L.M, de Vries, R.M.J.M, Ottmann, C. | | Deposit date: | 2017-01-26 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | Structural interface between LRRK2 and 14-3-3 protein.
Biochem. J., 474, 2017
|
|
3BA5
 
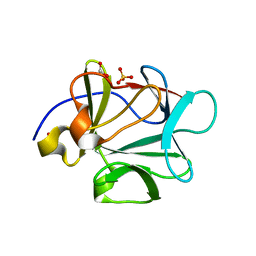 | |
3B4V
 
 | | X-Ray structure of Activin in complex with FSTL3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Inhibin beta A chain, ... | | Authors: | Thompson, T.B. | | Deposit date: | 2007-10-24 | | Release date: | 2008-09-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The structure of FSTL3.activin A complex. Differential binding of N-terminal domains influences follistatin-type antagonist specificity.
J.Biol.Chem., 283, 2008
|
|
3BA4
 
 | |
3BAH
 
 | |
5MPF
 
 | | Structural Basis of Gene Regulation by the Grainyhead Transcription Factor Superfamily | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*CP*CP*GP*GP*TP*TP*TP*T)-3'), Grainyhead-like protein 1 homolog | | Authors: | Ming, Q, Roske, Y, Schuetz, A, Walentin, K, Ibraimi, I, Schmidt-Ott, K.M, Heinemann, U. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family.
Nucleic Acids Res., 46, 2018
|
|
3B9U
 
 | |
2YPB
 
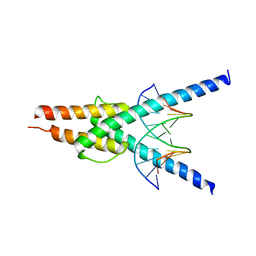 | | Structure of the SCL:E47 complex bound to DNA | | Descriptor: | EBOX FORWARD, EBOX REVERSE, T-CELL ACUTE LYMPHOCYTIC LEUKEMIA PROTEIN 1, ... | | Authors: | El Omari, K, Hoosdally, S.J, Tuladhar, K, Karia, D, Ponsele, E, Platonova, O, Vyas, P, Patient, R, Porcher, C, Mancini, E.J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural Basis for Lmo2-Driven Recruitment of the Scl:E47bHLH Heterodimer to Hematopoietic-Specific Transcriptional Targets.
Cell Rep., 4, 2013
|
|
7CVQ
 
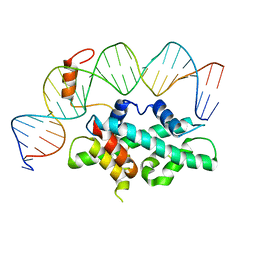 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB2/YC3 and FT CORE1 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-3 and Zinc finger protein CONSTANS, FT CORE1 DNA forward strand, FT CORE1 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
5MPI
 
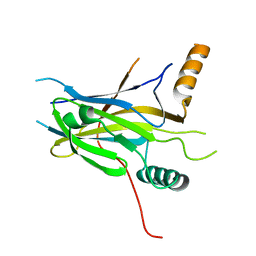 | | Structural Basis of Gene Regulation by the Grainyhead Transcription Factor Superfamily | | Descriptor: | Grainyhead-like protein 1 homolog | | Authors: | Ming, Q, Roske, Y, Schuetz, A, Walentin, K, Ibraimi, I, Schmidt-Ott, K.M, Heinemann, U. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family.
Nucleic Acids Res., 46, 2018
|
|
5EF6
 
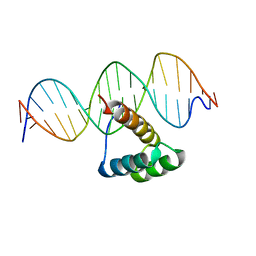 | | Structure of HOXB13 complex with methylated DNA | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-23 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
2YB8
 
 | | Crystal structure of Nurf55 in complex with Su(z)12 | | Descriptor: | POLYCOMB PROTEIN SU(Z)12, PROBABLE HISTONE-BINDING PROTEIN CAF1, SULFATE ION | | Authors: | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks.
Mol.Cell, 42, 2011
|
|
5MY9
 
 | | Crystal structure of human 14-3-3 sigma in complex with LRRK2 peptide pS935 | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stevers, L.M, de Vries, R.M.J.M, Ottmann, C. | | Deposit date: | 2017-01-26 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.327 Å) | | Cite: | Structural interface between LRRK2 and 14-3-3 protein.
Biochem. J., 474, 2017
|
|
6FIC
 
 | | Bivalent Inhibitor UNC4512 Bound to the TAF1 Bromodomain Tandem | | Descriptor: | 3-azanyl-2-[3-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[[3-azanyl-1-[[2-[[3-methyl-6-[4-methyl-3-(methylsulfonyl-$l^{2}-azanyl)cyclohexa-1,3,5-trien-1-yl]-[1,2,4]triazolo[4,3-b]pyridazin-8-yl]-$l^{2}-azanyl]-2-oxidanylidene-ethyl]amino]-1-oxidanylidene-propan-2-yl]amino]-2-oxidanylidene-ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]propanoylamino]-~{N}-[3-[[3-methyl-6-[4-methyl-3-(methylsulfonylamino)phenyl]-[1,2,4]triazolo[4,3-b]pyridazin-8-yl]amino]-3-oxidanylidene-propyl]propanamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Mathea, S, Suh, J.L, Salah, E, Tallant, C, Siejka, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, James, L.I, Frye, S.V, Knapp, S. | | Deposit date: | 2018-01-17 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Bivalent Inhibitor UNC4512 Bound to the TAF1 Bromodomain Tandem
To Be Published
|
|
6F07
 
 | | CBF3 Core Complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, Suppressor of kinetochore protein 1, ZINC ION | | Authors: | Leber, V, Singleton, M.R. | | Deposit date: | 2017-11-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for assembly of the CBF3 kinetochore complex.
EMBO J., 37, 2018
|
|
3BAG
 
 | |
7UYF
 
 | | Human PRMT5:MEP50 structure with Fragment 4 and MTA Bound | | Descriptor: | 4-methyl-1,5-naphthyridin-2-amine, 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, ... | | Authors: | Gunn, R.J, Lawson, J.D, Smith, C.R. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
