5QJV
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z281802060 | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-N-methylpyrimidin-2-amine, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QKB
 
 | | Ground-state model of NUDT5 and corresponding apo datasets for PanDDA analysis | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-12-07 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5QJF
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z52425517 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJU
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z906021418 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-2-(propan-2-yl)pyrimidine-4-carboxamide, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QKA
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z2377835233 | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-3-[(3R)-piperidin-3-yl]-1H-pyrazole-4-carboxamide, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5O81
 
 | |
5O9S
 
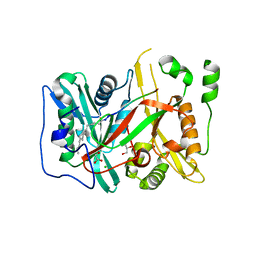 | | HsNMT1 in complex with CoA and Myristoylated-GKSNSKLK octapeptide | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Meinnel, T, Giglione, C. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern.
Nat. Chem. Biol., 14, 2018
|
|
5O91
 
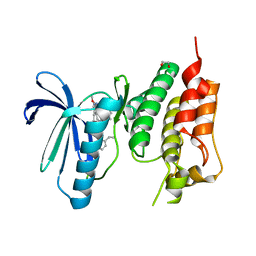 | | Crystal structure of human Mps1 (TTK) C604W mutant in complex with Cpd-5 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase TTK, ~{N}-(2,6-diethylphenyl)-8-[[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino]-1-methyl-4,5-dihydropyrazolo[4,3-h]quinazoline-3-carboxamide | | Authors: | Hiruma, Y, Joosten, R.P, Perrakis, A. | | Deposit date: | 2017-06-15 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Understanding inhibitor resistance in Mps1 kinase through novel biophysical assays and structures.
J. Biol. Chem., 292, 2017
|
|
5NWY
 
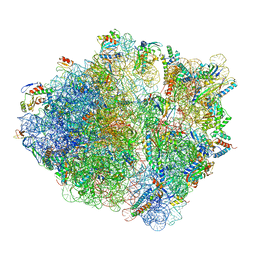 | | 2.9 A cryo-EM structure of VemP-stalled ribosome-nascent chain complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Cheng, J, Sohmen, D, Hedman, R, Berninghausen, O, von Heijne, G, Wilson, D.N, Beckmann, R. | | Deposit date: | 2017-05-08 | | Release date: | 2017-07-19 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The force-sensing peptide VemP employs extreme compaction and secondary structure formation to induce ribosomal stalling.
Elife, 6, 2017
|
|
5OCN
 
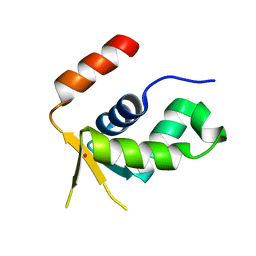 | | Crystal structure of the forkhead domain of human FOXN1 | | Descriptor: | Forkhead box protein N1, POTASSIUM ION | | Authors: | Newman, J.A, Aitkenhead, H, Pinkas, D.M, von Delft, F, Arrowsmith, C.H, Edwwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-07-03 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the forkhead domain of human FOXN1
To be published
|
|
5NSA
 
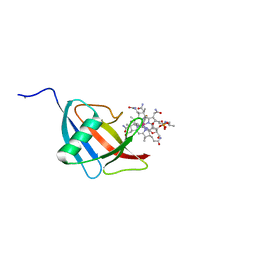 | | Beta domain of human transcobalamin bound to Co-beta-[2-(2,4-difluorophenyl)ethinyl]cobalamin | | Descriptor: | 1-ethynyl-2,4-difluorobenzene, CALCIUM ION, COBALAMIN, ... | | Authors: | Bloch, J.S, Ruetz, M, Krautler, B, Locher, K.P. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.273 Å) | | Cite: | Structure of the human transcobalamin beta domain in four distinct states.
PLoS ONE, 12, 2017
|
|
5NVM
 
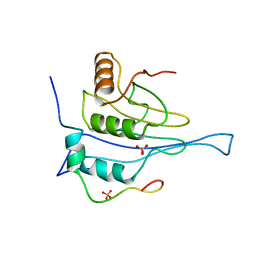 | |
5NSE
 
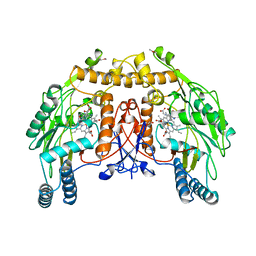 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, H4B-FREE, HYDROXY-ARG COMPLEX | | Descriptor: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-01-13 | | Release date: | 2002-05-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of the Heme Domain of Bovine Endothelial Nitric Oxide Synthase Complexed with Arginine Analogues
To be Published
|
|
5NVN
 
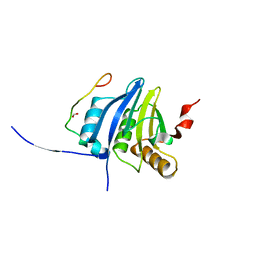 | | Crystal structure of the human 4EHP-4E-BP1 complex | | Descriptor: | Eukaryotic translation initiation factor 4E type 2, Eukaryotic translation initiation factor 4E-binding protein 1, FORMIC ACID | | Authors: | Peter, D, Sandmeir, F, Valkov, E. | | Deposit date: | 2017-05-04 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression.
Genes Dev., 31, 2017
|
|
5OC0
 
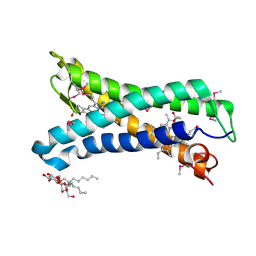 | | Structure of E. coli superoxide oxidase | | Descriptor: | Cytochrome b561, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lundgren, C.A.K, Sjostrand, D, Biner, O, Bennett, M, von Ballmoos, C, Hogbom, M. | | Deposit date: | 2017-06-29 | | Release date: | 2018-06-20 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Scavenging of superoxide by a membrane-bound superoxide oxidase.
Nat. Chem. Biol., 14, 2018
|
|
5OC9
 
 | | Crystal Structure of human TMEM16K / Anoctamin 10 | | Descriptor: | (2R)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-29 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
5O5E
 
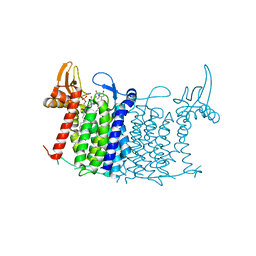 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) in complex with tunicamycin | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
5O2R
 
 | | Cryo-EM structure of the proline-rich antimicrobial peptide Api137 bound to the terminating ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2017-05-22 | | Release date: | 2017-07-26 | | Last modified: | 2019-02-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5O9V
 
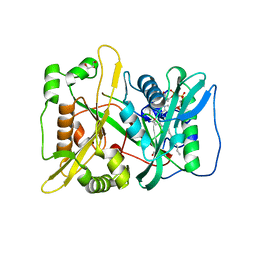 | | HsNMT1 in complex with CoA and Myristoylated-GGCFSKPK octapeptide | | Descriptor: | Apoptosis-inducing factor 3, CHLORIDE ION, COENZYME A, ... | | Authors: | Dian, C, Meinnel, T, Giglione, C. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern.
Nat. Chem. Biol., 14, 2018
|
|
5O7Z
 
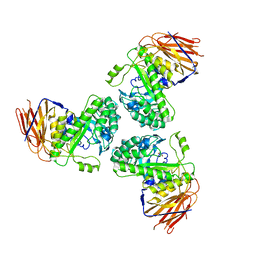 | |
5OFR
 
 | |
5OHZ
 
 | |
5OHT
 
 | | A GH31 family sulfoquinovosidase from E. coli in complex with aza-sugar inhibitor IFGSQ | | Descriptor: | CALCIUM ION, Sulfoquinovosidase, [(3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)piperidin-3-yl]methanesulfonic acid | | Authors: | Jin, Y, Williams, S.J, Goddard-Borger, E, Davies, G.J. | | Deposit date: | 2017-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Insights into the Function and Evolution of Sulfoquinovosidases.
ACS Cent Sci, 4, 2018
|
|
5QJ9
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z768399682 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-11-12 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJM
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z328695024 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
