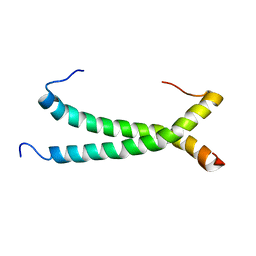
5GWM
 
 | |
8G4P
 
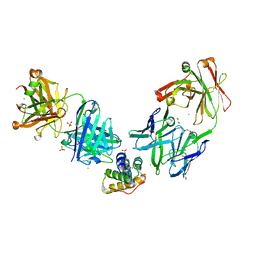 | | Crystal structure of the peanut allergen Ara h 2 bound by two neutralizing antibodies 13T1 and 13T5 | | Descriptor: | 1,2-ETHANEDIOL, 13T1 Fab light chain, 13T5 Fab heavy chain, ... | | Authors: | Pedersen, L.C, Mueller, G.A, Min, J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design of an Ara h 2 hypoallergen from conformational epitopes.
Clin Exp Allergy, 54, 2024
|
|
7OGB
 
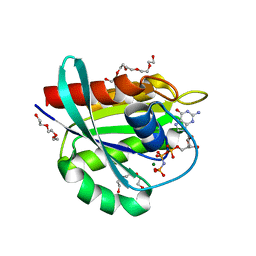 | | GTPase HRAS under 500 MPa pressure | | Descriptor: | GTPase HRas, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Colloc'h, N.C, Girard, E, Prange, T, Kalbitzer, H.R. | | Deposit date: | 2021-05-06 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Equilibria between conformational states of the Ras oncogene protein revealed by high pressure crystallography.
Chem Sci, 13, 2022
|
|
4X24
 
 | | Crystal structure of Vibrio cholerae 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, TRIETHYLENE GLYCOL | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-11-25 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site and remote contributions to catalysis in methylthioadenosine nucleosidases.
Biochemistry, 54, 2015
|
|
4X8F
 
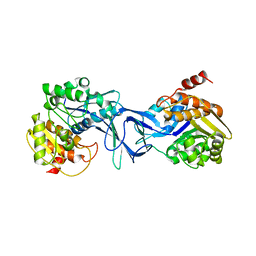 | | Vibrio cholerae O395 Ribokinase in apo form | | Descriptor: | Ribokinase | | Authors: | Paul, R, Patra, M.D, Sen, U. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of Apo and Ligand
Bound Vibrio choleraeRibokinase (Vc-RK):
Role of Monovalent Cation Induced Activation
and Structural Flexibility in Sugar
Phosphorylation
Adv Exp Med Biol., 842, 2015
|
|
4Z8A
 
 | | SH3-III of Drosophila Rim-binding protein bound to a Cacophony derived peptide | | Descriptor: | RIM-binding protein, isoform F, Voltage-dependent calcium channel type A subunit alpha-1 | | Authors: | Driller, J.H, Holton, N, Siebert, M, Boehme, A.M, Wahl, M.C, Sigrist, S.J, Loll, B. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | A high affinity RIM-binding protein/Aplip1 interaction prevents the formation of ectopic axonal active zones.
Elife, 4, 2015
|
|
4X23
 
 | |
5HNY
 
 | | Structural basis of backwards motion in kinesin-14: plus-end directed nKn669 in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2016-11-02 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
4X0G
 
 | | Structure of Bsg25A binding with DNA | | Descriptor: | ACETATE ION, Blastoderm-specific gene 25A, DNA (5'-D(*GP*TP*TP*CP*CP*AP*AP*TP*TP*GP*GP*AP*A)-3') | | Authors: | Ren, A. | | Deposit date: | 2014-11-21 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2065 Å) | | Cite: | Common and distinct DNA-binding and regulatory activities of the BEN-solo transcription factor family.
Genes Dev., 29, 2015
|
|
4WSF
 
 | | Falafel EVH1 domain bound to CENP-C FIM | | Descriptor: | Cenp-C, SULFATE ION, Serine/threonine-protein phosphatase 4 regulatory subunit 3 | | Authors: | Lefevre, S.R, Singleton, M.R. | | Deposit date: | 2014-10-27 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Centromeric binding and activity of Protein Phosphatase 4.
Nat Commun, 6, 2015
|
|
4WSI
 
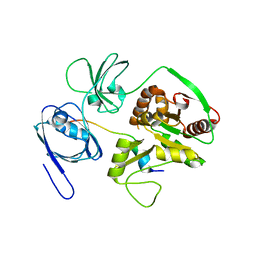 | | Crystal Structure of PALS1/Crb complex | | Descriptor: | MAGUK p55 subfamily member 5, Protein crumbs | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of Crumbs tail in complex with the PALS1 PDZ-SH3-GK tandem reveals a highly specific assembly mechanism for the apical Crumbs complex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3EIN
 
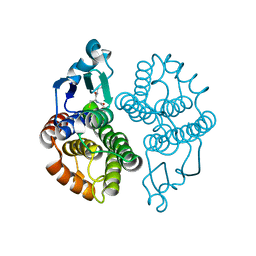 | | Delta class GST | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 1-1 | | Authors: | Feil, S.C. | | Deposit date: | 2008-09-17 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.126 Å) | | Cite: | Probing insect detoxification systems
To be Published
|
|
3EKU
 
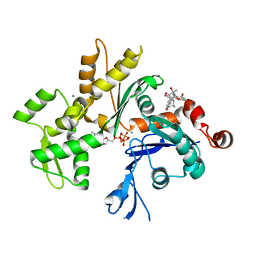 | | Crystal Structure of Monomeric Actin bound to Cytochalasin D | | Descriptor: | (3S,3aR,4S,6S,6aR,7E,10S,12R,13E,15R,15aR)-3-benzyl-6,12-dihydroxy-4,10,12-trimethyl-5-methylidene-1,11-dioxo-2,3,3a,4,5,6,6a,9,10,11,12,15-dodecahydro-1H-cycloundeca[d]isoindol-15-yl acetate, ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, ... | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
4Z89
 
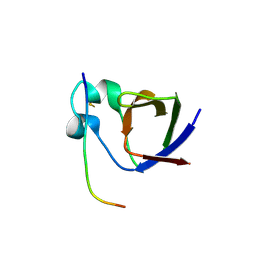 | | SH3-II of Drosophila Rim-binding protein bound to a Cacophony derived peptide | | Descriptor: | CALCIUM ION, RIM-binding protein, isoform F, ... | | Authors: | Driller, J.H, Holton, N, Siebert, M, Boehme, M.A, Wahl, M.C, Sigrist, S.J, Loll, B. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A high affinity RIM-binding protein/Aplip1 interaction prevents the formation of ectopic axonal active zones.
Elife, 4, 2015
|
|
4Z88
 
 | | SH3-II of Drosophila Rim-binding protein with Aplip1 peptide | | Descriptor: | JNK-interacting protein 1, PHOSPHATE ION, RIM-binding protein, ... | | Authors: | Driller, J.H, Holton, N, Siebert, M, Boehme, M.A, Wahl, M.C, Sigrist, S.J, Loll, B. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A high affinity RIM-binding protein/Aplip1 interaction prevents the formation of ectopic axonal active zones.
Elife, 4, 2015
|
|
4ZLR
 
 | | Structure of the Brat-NHL domain bound to consensus RNA motif | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Brain tumor protein, PENTAETHYLENE GLYCOL, ... | | Authors: | Jakob, L, Treiber, N, Treiber, T, Stotz, M, Loedige, I, Meister, G. | | Deposit date: | 2015-05-01 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the NHL Domain in Complex with RNA Reveals the Molecular Basis of Drosophila Brain-Tumor-Mediated Gene Regulation.
Cell Rep, 13, 2015
|
|
3GEC
 
 | |
3GMJ
 
 | | Crystal structure of MAD MH2 domain | | Descriptor: | Protein mothers against dpp | | Authors: | Wu, J.W, Wang, C. | | Deposit date: | 2009-03-14 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the MH2 domain of Drosophila Mad
SCI.CHINA, SER.C: LIFE SCI., 52, 2009
|
|
3EL2
 
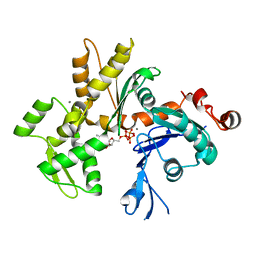 | | Crystal Structure of Monomeric Actin Bound to Ca-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
3E9T
 
 | | Crystal structure of Apo-form Calx CBD1 domain | | Descriptor: | CALCIUM ION, Na/Ca exchange protein | | Authors: | Wu, M, Zheng, L. | | Deposit date: | 2008-08-23 | | Release date: | 2009-09-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of progressive Ca2+ binding states of the Ca2+ sensor Ca2+ binding domain 1 (CBD1) from the CALX Na+/Ca2+ exchanger reveal incremental conformational transitions.
J.Biol.Chem., 285, 2010
|
|
3GN4
 
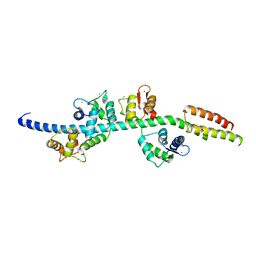 | | Myosin lever arm | | Descriptor: | CALCIUM ION, Calmodulin, MAGNESIUM ION, ... | | Authors: | Mukherjea, M, Llinas, P, Kim, H, Travaglia, M, Safer, D, Zong, A.B, Menetrey, J, Franzini-Armstrong, C, Selvin, P.R, Houdusse, A, Sweeney, H.L. | | Deposit date: | 2009-03-16 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Myosin VI dimerization triggers an unfolding of a three-helix bundle in order to extend its reach
Mol.Cell, 35, 2009
|
|
7KEI
 
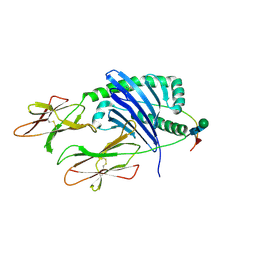 | | DQA1*01:02/DQB1*06:02 in complex with a hemagglutinin peptide from the H1N1 pandemic flu virus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HA peptide from 2009 H1N1 pandemic flu virus., ... | | Authors: | Birtley, J.R, Stern, L.J, Mellins, E.D, Jiang, W. | | Deposit date: | 2020-10-10 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of DQA1*01:02/DQB1*06:02 in complex with a flu peptide.
To Be Published
|
|
7LFM
 
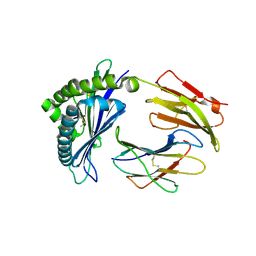 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, VAL MUTANT, TRICLINIC CELL, REFINED AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFL
 
 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, VAL MUTANT, MONOCLINIC CELL, REFINED AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFK
 
 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, THR MUTANT, REFINED AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
