2Q9N
 
 | | 4-Substituted Trinems as Broad Spectrum-Lactamase Inhibitors: Structure-based Design, Synthesis and Biological Activity | | Descriptor: | (1S,4R,7AR)-4-BUTOXY-1-[(1R)-1-FORMYLPROPYL]-2,4,5,6,7,7A-HEXAHYDRO-1H-ISOINDOLE-3-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Plantan, I, Selic, L, Mesar, T, Stefanic Anderluh, P, Oblak, M, Prezelj, A, Hesse, L, Andrejasic, M, Vilar, M, Turk, D, Kocijan, A, Prevec, T, Vilfan, G, Kocjan, D, Copar, A, Urleb, U, Solmajer, T. | | Deposit date: | 2007-06-13 | | Release date: | 2007-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 4-Substituted Trinems as Broad Spectrum beta-Lactamase Inhibitors: Structure-Based Design, Synthesis, and Biological Activity
J.Med.Chem., 50, 2007
|
|
7AN4
 
 | |
7ANJ
 
 | |
7ANI
 
 | |
7ANG
 
 | |
7ANC
 
 | |
4JMG
 
 | |
2P9A
 
 | | E. coli methionine aminopeptidase dimetalated with inhibitor YE6 | | Descriptor: | 5-(2-chlorophenyl)furan-2-carbohydrazide, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q. | | Deposit date: | 2007-03-24 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of Monometalated Methionine Aminopeptidase: Inhibitor Discovery and Crystallographic Analysis.
J.Med.Chem., 50, 2007
|
|
2P99
 
 | | E. coli methionine aminopeptidase monometalated with inhibitor YE6 | | Descriptor: | 5-(2-chlorophenyl)furan-2-carbohydrazide, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q. | | Deposit date: | 2007-03-24 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of Monometalated Methionine Aminopeptidase: Inhibitor Discovery and Crystallographic Analysis.
J.Med.Chem., 50, 2007
|
|
3OK5
 
 | |
3OJ1
 
 | |
1RGY
 
 | | Citrobacter freundii GN346 Class C beta-Lactamase Complexed with Transition-State Analog of Cefotaxime | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, beta-lactamase, {[(2E)-2-(2-AMINO-1,3-THIAZOL-4-YL)-2-(METHOXYIMINO)ETHANOYL]AMINO}METHYLPHOSPHONIC ACID | | Authors: | Nukaga, M, Kumar, S, Nukaga, K, Pratt, R.F, Knox, J.R. | | Deposit date: | 2003-11-13 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Hydrolysis of third-generation cephalosporins by class C beta-lactamases. Structures of a transition state analog of cefotoxamine in wild-type and extended spectrum enzymes.
J.Biol.Chem., 279, 2004
|
|
1RGZ
 
 | | Enterobacter cloacae GC1 Class C beta-Lactamase Complexed with Transition-State Analog of Cefotaxime | | Descriptor: | GLYCEROL, class C beta-lactamase, {[(2E)-2-(2-AMINO-1,3-THIAZOL-4-YL)-2-(METHOXYIMINO)ETHANOYL]AMINO}METHYLPHOSPHONIC ACID | | Authors: | Nukaga, M, Kumar, S, Nukaga, K, Pratt, R.F, Knox, J.R. | | Deposit date: | 2003-11-13 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Hydrolysis of third-generation cephalosporins by class C beta-lactamases. Structures of a transition state analog of cefotoxamine in wild-type and extended spectrum enzymes.
J.Biol.Chem., 279, 2004
|
|
8DOH
 
 | | Dehaloperoxidase B in complex with Bisphenol F | | Descriptor: | 4,4'-methylenediphenol, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serrano, V.S, Yun, D, Ghiladi, R. | | Deposit date: | 2022-07-13 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Oxidation of bisphenol A (BPA) and related compounds by the multifunctional catalytic globin dehaloperoxidase.
J.Inorg.Biochem., 238, 2023
|
|
8DOG
 
 | | Dehaloperoxidase B in complex with Bisphenol E | | Descriptor: | 4-[1-(4-hydroxyphenyl)ethyl]phenol, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serrano, V.S, Yun, D, Ghiladi, R. | | Deposit date: | 2022-07-13 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Oxidation of bisphenol A (BPA) and related compounds by the multifunctional catalytic globin dehaloperoxidase.
J.Inorg.Biochem., 238, 2023
|
|
8DOJ
 
 | | Dehaloperoxidase B in complex with 3,3'-Biphenol | | Descriptor: | (1P)-[1,1'-biphenyl]-3,3'-diol, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serrano, V.S, Yun, D, Ghiladi, R. | | Deposit date: | 2022-07-13 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oxidation of bisphenol A (BPA) and related compounds by the multifunctional catalytic globin dehaloperoxidase.
J.Inorg.Biochem., 238, 2023
|
|
8DOI
 
 | | Dehaloperoxidase B in complex with 2,2'-Biphenol | | Descriptor: | 2-(2-hydroxyphenyl)phenol, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | de Serrano, V.S, Yun, D, Ghiladi, R. | | Deposit date: | 2022-07-13 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Oxidation of bisphenol A (BPA) and related compounds by the multifunctional catalytic globin dehaloperoxidase.
J.Inorg.Biochem., 238, 2023
|
|
4JMH
 
 | |
2V65
 
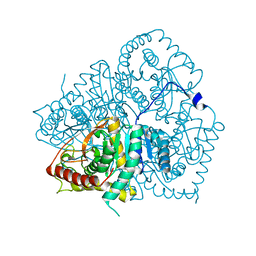 | | Apo LDH from the psychrophile C. gunnari | | Descriptor: | L-LACTATE DEHYDROGENASE A CHAIN | | Authors: | Coquelle, N, Madern, D, Vellieux, F. | | Deposit date: | 2007-07-13 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Activity, Stability and Structural Studies of Lactate Dehydrogenases Adapted to Extreme Thermal Environments.
J.Mol.Biol., 374, 2007
|
|
4L4X
 
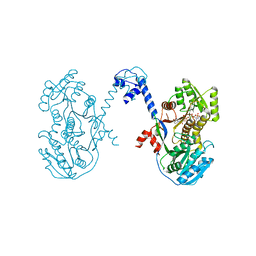 | | An A2-type ketoreductase from a modular polyketide synthase | | Descriptor: | AmphI, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, J, Keatinge-Clay, A.T. | | Deposit date: | 2013-06-10 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Studies of an A2-Type Modular Polyketide Synthase Ketoreductase Reveal Features Controlling alpha-Substituent Stereochemistry.
Acs Chem.Biol., 8, 2013
|
|
8S6N
 
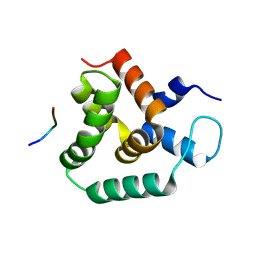 | | Structure of MLLE3 in complex with PAMPL1 | | Descriptor: | PAMPL1, RNA-binding protein RRM4 | | Authors: | Devan, S, Shanmugasundaram, S, Muentjes, K, Smits, S.H, Altegoer, F, Feldbruegge, M. | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | Deciphering the RNA-binding protein network during endosomal mRNA transport
To Be Published
|
|
8S6O
 
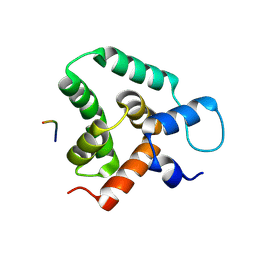 | | Structure of MLLE3 in complex with PAMPL2 | | Descriptor: | PAMPL2, RNA-binding protein RRM4 | | Authors: | Devan, S, Shanmugasundaram, S, Muentjes, K, Smits, S.H, Altegoer, F, Feldbruegge, M. | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Deciphering the RNA-binding protein network during endosomal mRNA transport
To Be Published
|
|
8SWM
 
 | |
5OGL
 
 | | Structure of bacterial oligosaccharyltransferase PglB in complex with an acceptor peptide and an lipid-linked oligosaccharide analog | | Descriptor: | MANGANESE (II) ION, SODIUM ION, Substrate mimicking peptide, ... | | Authors: | Napiorkowska, M, Boilevin, J, Sovdat, T, Darbre, T, Reymond, J.-L, Aebi, M, Locher, K.P. | | Deposit date: | 2017-07-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6NSJ
 
 | | CryoEM structure of Helicobacter pylori urea channel in closed state | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, Acid-activated urea channel | | Authors: | Cui, Y.X, Zhou, K, Strugatsky, D, Wen, Y, Sachs, G, Munson, K, Zhou, Z.H. | | Deposit date: | 2019-01-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | pH-dependent gating mechanism of theHelicobacter pyloriurea channel revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
