2VAX
 
 | | Crystal structure of deacetylcephalosporin C acetyltransferase (Cephalosporin C-soak) | | Descriptor: | 4-(3-ACETOXYMETHYL-2-CARBOXY-8-OXO-5-THIA-1-AZA-BICYCLO[4.2.0]OCT-2-EN-7-YLCARBAMOYL)-1-CARBOXY-BUTYL-AMMONIUM, ACETATE ION, ACETYL-COA--DEACETYLCEPHALOSPORIN C ACETYLTRANSFERASE | | Authors: | Lejon, S, Ellis, J, Valegard, K. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Last Step in Cephalosporin C Formation Revealed: Crystal Structures of Deacetylcephalosporin C Acetyltransferase from Acremonium Chrysogenum in Complexes with Reaction Intermediates.
J.Mol.Biol., 377, 2008
|
|
4HHP
 
 | | Crystal structure of triosephosphate isomerase from trypanosoma cruzi, mutant e105d | | Descriptor: | GLYCEROL, SULFATE ION, Triosephosphate isomerase, ... | | Authors: | Hernandez-Santoyo, A, Aguirre-Fuentes, Y, Torres-Larios, A, Gomez-Puyou, A, De Gomez-Puyou, M.T. | | Deposit date: | 2012-10-10 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Different contribution of conserved amino acids to the global properties of triosephosphate isomerases.
Proteins, 82, 2014
|
|
9GFJ
 
 | | Crystal structure of ASO binding Fab fragment with ASO143 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1~{S},3~{R},4~{R},7~{S})-7-[[(1~{R},3~{R},4~{R},7~{S})-7-[[(1~{R},3~{R},4~{R},7~{S})-3-(6-aminopurin-9-yl)-7-[[(1~{R},3~{R},4~{R},7~{S})-3-(4-azanyl-5-methyl-2-oxidanylidene-pyrimidin-1-yl)-7-oxidanyl-2,5-dioxabicyclo[2.2.1]heptan-1-yl]methoxy-sulfanyl-phosphoryl]oxy-2,5-dioxabicyclo[2.2.1]heptan-1-yl]methoxy-sulfanyl-phosphoryl]oxy-3-(4-azanyl-5-methyl-2-oxidanylidene-pyrimidin-1-yl)-2,5-dioxabicyclo[2.2.1]heptan-1-yl]methoxy-sulfanyl-phosphoryl]oxy-1-(hydroxymethyl)-2,5-dioxabicyclo[2.2.1]heptan-3-yl]-5-methyl-pyrimidine-2,4-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hsia, H.-E, Zanini, C, Simonneau, C, Fraidling, J, Kraft, T, Mayer, K, Sommer, A, Indlekofer, A, Wirht, T, Benz, J, Georges, G, Langer, L.M, Gassner, C, Larraillet, V, Manso, M, Ravn, J, Hofer, K, Emrich, T, Niewoehner, J, Schumacher, F, Brinkmann, U. | | Deposit date: | 2024-08-09 | | Release date: | 2025-08-20 | | Last modified: | 2025-11-12 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Improved targeted delivery of antisense oligonucleotide with an antibody mask.
Nucleic Acids Res., 53, 2025
|
|
9GFD
 
 | | Crystal structure of ASO binding Fab fragment with ASO139 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1~{S},3~{R},4~{R},7~{S})-7-[[(1~{R},3~{R},4~{R},7~{S})-7-[[(1~{R},3~{R},4~{R},7~{S})-3-(6-aminopurin-9-yl)-7-[[(2~{R},3~{S},5~{R})-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3-oxidanyl-oxolan-2-yl]methoxy-sulfanyl-phosphoryl]oxy-2,5-dioxabicyclo[2.2.1]heptan-1-yl]methoxy-sulfanyl-phosphoryl]oxy-3-(4-azanyl-5-methyl-2-oxidanylidene-pyrimidin-1-yl)-2,5-dioxabicyclo[2.2.1]heptan-1-yl]methoxy-sulfanyl-phosphoryl]oxy-1-(hydroxymethyl)-2,5-dioxabicyclo[2.2.1]heptan-3-yl]-5-methyl-pyrimidine-2,4-dione, Fab Fragment light chain, ... | | Authors: | Hsia, H.-E, Zanini, C, Simonneau, C, Fraidling, J, Kraft, T, Mayer, K, Sommer, A, Indlekofer, A, Wirth, T, Benz, J, Geroges, G, Langer, M.L, Gassner, C, Larraillet, V, Manso, M, Ravn, J, Hofer, K, Emrich, T, Niewoehner, J, Schumacher, F, Brinkmann, U. | | Deposit date: | 2024-08-09 | | Release date: | 2025-08-20 | | Last modified: | 2025-11-12 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Improved targeted delivery of antisense oligonucleotide with an antibody mask.
Nucleic Acids Res., 53, 2025
|
|
4KUM
 
 | | Structure of LSD1-CoREST-Tetrahydrofolate complex | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Calcutt, M.W, Newcomer, M.E, Wagner, C. | | Deposit date: | 2013-05-22 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of the histone lysine specific demethylase LSD1 complexed with tetrahydrofolate.
Protein Sci., 23, 2014
|
|
4L9O
 
 | | Crystal Structure of the Sec13-Sec16 blade-inserted complex from Pichia pastoris | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | McMahon, C, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2013-06-18 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sec16 influences transitional ER sites by regulating rather than organizing COPII.
Mol Biol Cell, 24, 2013
|
|
6N1K
 
 | |
2VI4
 
 | |
4O6B
 
 | |
9L8R
 
 | | Dihydroxyacid dehydratase (DHAD) | | Descriptor: | (2~{R})-7-fluoranyl-2-oxidanyl-3-oxidanylidene-4-prop-2-ynyl-1,4-benzoxazine-2-carboxylic acid, Dihydroxy-acid dehydratase, chloroplastic, ... | | Authors: | Zhou, J, Liu, D. | | Deposit date: | 2024-12-28 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The mutant structure of DHAD
To Be Published
|
|
1NFU
 
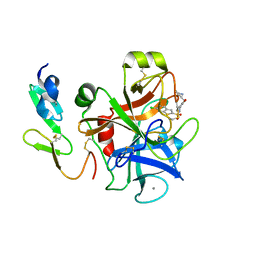 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR XA COMPLEXED WITH RPR132747 | | Descriptor: | 3-({4-[(6-CHLORO-1-BENZOTHIEN-2-YL)SULFONYL]-2-OXOPIPERAZIN-1-YL}METHYL)BENZENECARBOXIMIDAMIDE, CALCIUM ION, COAGULATION FACTOR XA, ... | | Authors: | Maignan, S, Guilloteau, J.P. | | Deposit date: | 2002-12-16 | | Release date: | 2003-02-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular structures of human Factor Xa complexed with ketopiperazine inhibitors: preference for a neutral group in the S1 pocket.
J.Med.Chem., 46, 2003
|
|
5AR2
 
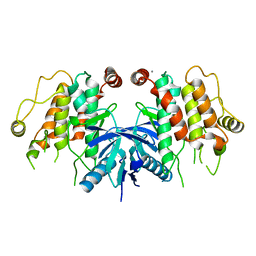 | | RIP2 Kinase Catalytic Domain (1 - 310) | | Descriptor: | CALCIUM ION, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-23 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
1NFY
 
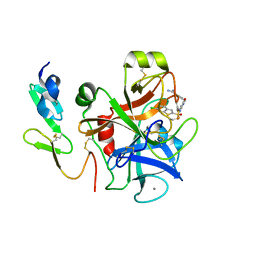 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR XA COMPLEXED WITH RPR200095 | | Descriptor: | 4-({4-[(6-CHLORO-1-BENZOTHIEN-2-YL)SULFONYL]-2-OXOPIPERAZIN-1-YL}METHYL)BENZENECARBOXIMIDAMIDE, CALCIUM ION, Coagulation factor XA, ... | | Authors: | Maignan, S, Guilloteau, J.P. | | Deposit date: | 2002-12-16 | | Release date: | 2003-02-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular structures of human Factor Xa complexed with ketopiperazine inhibitors: preference for a neutral group in the S1 pocket.
J.Med.Chem., 46, 2003
|
|
2VF0
 
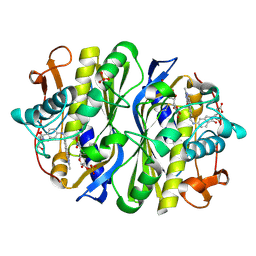 | | CRYSTAL STRUCTURE OF THE THYMIDYLATE SYNTHASE K48Q COMPLEXED WITH 5NO2DUMP AND BW1843U89 | | Descriptor: | 2'-DEOXY-5-NITROURIDINE 5'-MONOPHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, SULFATE ION, ... | | Authors: | Sotelo-Mundo, R.R, Arreola, R, Maley, F, Montfort, W.R. | | Deposit date: | 2007-10-27 | | Release date: | 2007-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Role of an Invariant Lysine Residue in Folate Binding on Escherichia Coli Thymidylate Synthase: Calorimetric and Crystallographic Analysis of the K48Q Mutant.
Int.J.Biochem.Cell Biol., 40, 2008
|
|
4GV2
 
 | | Human ARTD3 (PARP3) - Catalytic domain in complex with inhibitor ME0354 | | Descriptor: | 3-(4-oxo-3,4-dihydroquinazolin-2-yl)-N-[(1R)-1-(pyridin-2-yl)ethyl]propanamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 3 | | Authors: | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | Deposit date: | 2012-08-30 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | PARP Inhibitor with Selectivity Toward ADP-Ribosyltransferase ARTD3/PARP3
Acs Chem.Biol., 8, 2013
|
|
1NC3
 
 | | Crystal structure of E. coli MTA/AdoHcy nucleosidase complexed with formycin A (FMA) | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2002-12-04 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Escherichia coli 5'-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis.
J.Biol.Chem., 278, 2003
|
|
5VD5
 
 | |
3BHV
 
 | | Structure of phosphorylated Thr160 CDK2/cyclin A in complex with the inhibitor variolin B | | Descriptor: | 9-amino-5-(2-aminopyrimidin-4-yl)pyrido[3',2':4,5]pyrrolo[1,2-c]pyrimidin-4-ol, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Echalier, A, Bettayeb, K, Ferandin, Y, Lozach, O, Clement, M, Valette, A, Liger, F, Marquet, B, Morris, J.C, Endicott, J.A, Joseph, B, Meijer, L. | | Deposit date: | 2007-11-29 | | Release date: | 2008-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Meriolins, a new class of cell death inducing kinase inhibitors with enhanced selectivity for cyclin-dependent kinases
Cancer Res., 67, 2007
|
|
1I6S
 
 | | T4 LYSOZYME MUTANT C54T/C97A/N101A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Kovall, R.A, Baldwin, E.P, Matthews, B.W. | | Deposit date: | 2001-03-04 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
7C52
 
 | | Co-crystal structure of a photosynthetic LH1-RC in complex with electron donor HiPIP | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Yu, L.-J, Wang-Otomo, Z.-Y. | | Deposit date: | 2020-05-18 | | Release date: | 2021-03-03 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of a photosynthetic LH1-RC in complex with its electron donor HiPIP.
Nat Commun, 12, 2021
|
|
2AL6
 
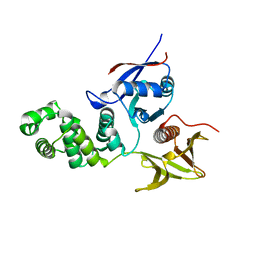 | | FERM domain of Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Ceccarelli, D.F, Song, H.K, Poy, F, Schaller, M.D, Eck, M.J. | | Deposit date: | 2005-08-04 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the FERM Domain of Focal Adhesion Kinase
J.Biol.Chem., 281, 2006
|
|
3DAY
 
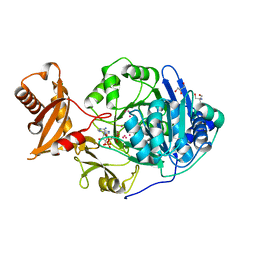 | | Crystal structure of human acyl-CoA synthetase medium-chain family member 2A (L64P mutation) in complex with AMP-CPP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acyl-coenzyme A synthetase ACSM2A, mitochondrial precursor, ... | | Authors: | Pilka, E.S, Kochan, G.T, Yue, W.W, Bhatia, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wikstrom, M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural snapshots for the conformation-dependent catalysis by human medium-chain acyl-coenzyme A synthetase ACSM2A.
J.Mol.Biol., 388, 2009
|
|
7GGJ
 
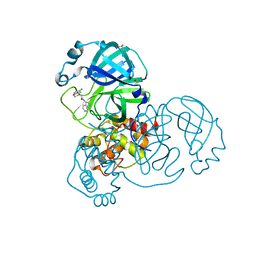 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BEN-DND-4f474d93-1 (Mpro-x12659) | | Descriptor: | (4R)-6-chloro-N-(2,7-naphthyridin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
2FC8
 
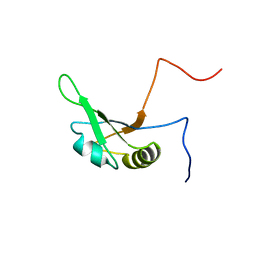 | | Solution structure of the RRM_1 domain of NCL protein | | Descriptor: | NCL protein | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-12 | | Release date: | 2006-06-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM_1 domain of NCL protein
To be published
|
|
4D7Q
 
 | | Crystal structure of a chimeric protein with the Sec7 domain of Legionella pneumophila RalF and the capping domain of Rickettsia prowazekii RalF | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Folly-Klan, M, Sancerne, B, Alix, E, Roy, C.R, Cherfils, J, Campanacci, V. | | Deposit date: | 2014-11-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | On the Use of Legionella/Rickettsia Chimeras to Investigate the Structure and Regulation of Rickettsia Effector Ralf.
J.Struct.Biol., 189, 2015
|
|
