7SXY
 
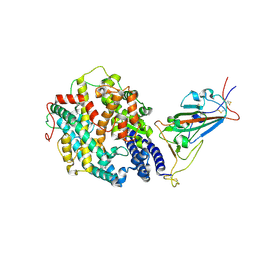 | | Cryo-EM structure of the SARS-CoV-2 D614G mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SY3
 
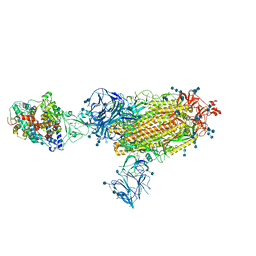 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K mutant spike protein ectodomain bound to human ACE2 ectodomain (global refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SY8
 
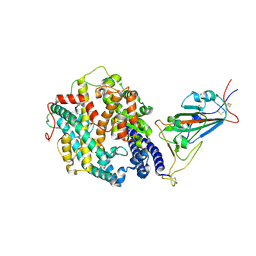 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K,K417T mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SXZ
 
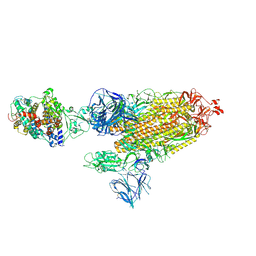 | | Cryo-EM structure of the SARS-CoV-2 D614G,L452R mutant spike protein ectodomain bound to human ACE2 ectodomain (global refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SXX
 
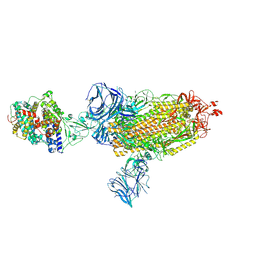 | | Cryo-EM structure of the SARS-CoV-2 D614G mutant spike protein ectodomain bound to human ACE2 ectodomain (global refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
5CO7
 
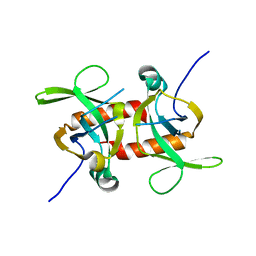 | | E. coli MazF form III | | Descriptor: | Endoribonuclease MazF | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-19 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.489 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
7SVR
 
 | |
7T7J
 
 | |
7SV7
 
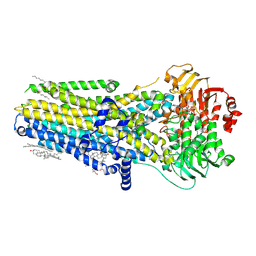 | |
4NSX
 
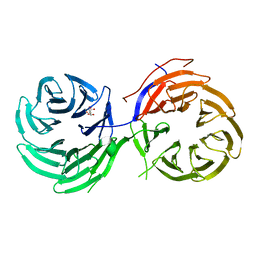 | | Crystal Structure of the Utp21 tandem WD Domain | | Descriptor: | CITRATE ANION, U3 small nucleolar RNA-associated protein 21 | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Utp21 Tandem WD Domain Provides Insight into the Organization of the UTPB Complex Involved in Ribosome Synthesis
Plos One, 9, 2014
|
|
7SVD
 
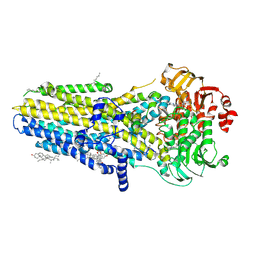 | |
6XBM
 
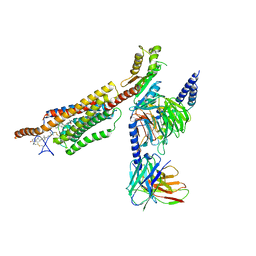 | | Structure of human SMO-Gi complex with 24(S),25-EC | | Descriptor: | 17-[3-(3,3-DIMETHYL-OXIRANYL)-1-METHYL-PROPYL]-10,13-DIMETHYL-2,3,4,7,8,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYC LOPENTA[A]PHENANTHREN-3-OL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qi, X, Long, T, Li, X. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Sterols in an intramolecular channel of Smoothened mediate Hedgehog signaling.
Nat.Chem.Biol., 16, 2020
|
|
6XO3
 
 | | ScoE with alpha-ketoglutarate in an off-site | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, FE (II) ION, ... | | Authors: | Jonnalagadda, R, Drennan, C.L. | | Deposit date: | 2020-07-06 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and crystallographic investigations into isonitrile formation by a nonheme iron-dependent oxidase/decarboxylase.
J.Biol.Chem., 296, 2021
|
|
7TDF
 
 | | AtTPC1 D454N with 1 mM EDTA state I | | Descriptor: | Two pore calcium channel protein 1 | | Authors: | Dickinson, M.S, Stroud, R.M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis of multistep voltage activation in plant two-pore channel 1.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TBG
 
 | | AtTPC1 D454N with 1 mM Ca2+ | | Descriptor: | CALCIUM ION, SODIUM ION, Two pore calcium channel protein 1 | | Authors: | Dickinson, M.S, Stroud, R.M. | | Deposit date: | 2021-12-22 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of multistep voltage activation in plant two-pore channel 1.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TDM
 
 | |
7TDN
 
 | |
7ST6
 
 | |
7SS9
 
 | | Late translocation intermediate with EF-G partially dissociated (Structure V) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Carbone, C.E, Loveland, A.B, Gamper, H.B, Hou, Y, Korostelev, A.A. | | Deposit date: | 2021-11-10 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Time-resolved cryo-EM visualizes ribosomal translocation with EF-G and GTP.
Nat Commun, 12, 2021
|
|
7TAP
 
 | | Cryo-EM structure of archazolid A bound to yeast VO V-ATPase | | Descriptor: | Archazolid A, V-type proton ATPase subunit a, vacuolar isoform, ... | | Authors: | Keon, K.A, Rubinstein, J.L, Benlekbir, S, Kirsch, S.H, Muller, R. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM of the Yeast V O Complex Reveals Distinct Binding Sites for Macrolide V-ATPase Inhibitors.
Acs Chem.Biol., 17, 2022
|
|
7ST7
 
 | |
7ST2
 
 | |
7SSO
 
 | |
7SSW
 
 | |
7SSD
 
 | |
