4C4C
 
 | | Michaelis complex of Hypocrea jecorina CEL7A E217Q mutant with cellononaose spanning the active site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, COBALT (II) ION, ... | | Authors: | Haddad-Momeni, M, Sandgren, M, Stahlberg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Mechanism of Cellulose Hydrolysis by a Two-Step, Retaining Cellobiohydrolase Elucidated by Structural and Transition Path Sampling Studies.
J.Am.Chem.Soc., 136, 2014
|
|
4BQ6
 
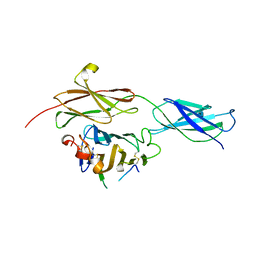 | | Crystal structure of the RGMB-NEO1 complex form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEOGENIN, RGM DOMAIN FAMILY MEMBER B | | Authors: | Bell, C.H, Healey, E, van Erp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
6AZ0
 
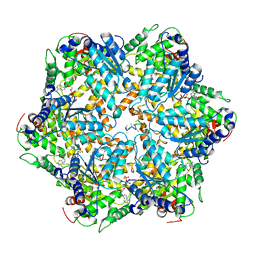 | | Mitochondrial ATPase Protease YME1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Puchades, C, Rampello, A.J, Shin, M, Giuliano, C, Wiseman, R.L, Glynn, S.E, Lander, G.C. | | Deposit date: | 2017-09-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing.
Science, 358, 2017
|
|
4BMK
 
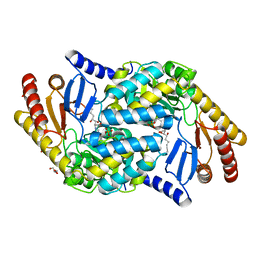 | | Serine Palmitoyltransferase K265A from S. paucimobilis with bound PLP- Myriocin Aldimine | | Descriptor: | Decarboxylated Myriocin, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Wadsworth, J.M, Clarke, D.J, McMahon, S.A, Beattie, A.E, Lowther, J, Dunn, T.M, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2013-05-09 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The Chemical Basis of Serine Palmitoyltransferase Inhibition by Myriocin.
J.Am.Chem.Soc., 135, 2013
|
|
4BVU
 
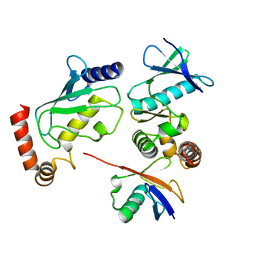 | | Structure of Shigella effector OspG in complex with host UbcH5c- Ubiquitin conjugate | | Descriptor: | PROTEIN KINASE OSPG, UBIQUITIN, UBIQUITIN-CONJUGATING ENZYME E2 D3 | | Authors: | Pruneda, J.N, LeTrong, I, Stenkamp, R.E, Klevit, R.E, Brzovic, P.S. | | Deposit date: | 2013-06-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | E2~Ub Conjugates Regulate the Kinase Activity of Shigella Effector Ospg During Pathogenesis.
Embo J., 33, 2014
|
|
4C0A
 
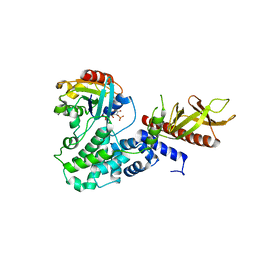 | | Arf1(Delta1-17)in complex with BRAG2 Sec7-PH domain | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, IQ MOTIF AND SEC7 DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Aizel, K, Biou, V, Navaza, J, Duarte, L, Campanacci, V, Cherfils, J, Zeghouf, M. | | Deposit date: | 2013-08-01 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Integrated conformational and lipid-sensing regulation of endosomal ArfGEF BRAG2.
PLoS Biol., 11, 2013
|
|
8W88
 
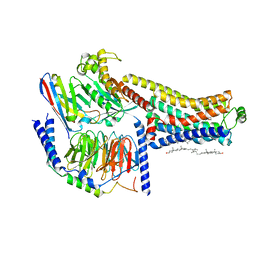 | | Cryo-EM structure of the SEP363856-bound TAAR1-Gs complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W8A
 
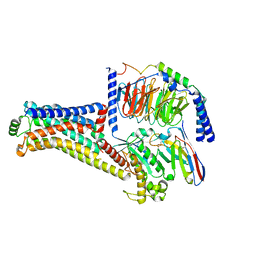 | | Cryo-EM structure of the RO5256390-TAAR1 complex | | Descriptor: | (4S)-4-[(2S)-2-phenylbutyl]-1,3-oxazolidin-2-imine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
4AF0
 
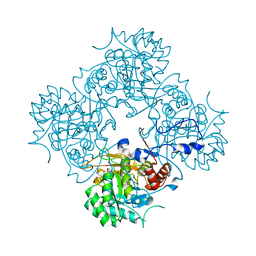 | | Crystal structure of cryptococcal inosine monophosphate dehydrogenase | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID, MYCOPHENOLIC ACID, ... | | Authors: | Valkov, E, Stamp, A, Morrow, C.A, Kobe, B, Fraser, J.A. | | Deposit date: | 2012-01-15 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De Novo GTP Biosynthesis is Critical for Virulence of the Fungal Pathogen Cryptococcus Neoformans
Plos Pathog., 8, 2012
|
|
8W89
 
 | | Cryo-EM structure of the PEA-bound TAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
6BGP
 
 | |
6BJD
 
 | | Crystal Structure of Human Calpain-3 Protease Core in Complex with E-64 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Calpain-3, ... | | Authors: | Ye, Q, Campbell, R.L, Davies, P.L. | | Deposit date: | 2017-11-06 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of human calpain-3 protease core with and without bound inhibitor reveal mechanisms of calpain activation.
J. Biol. Chem., 293, 2018
|
|
4CBL
 
 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
4CBH
 
 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
4A6W
 
 | | X-ray structures of oxazole hydroxamate EcMetAp-Mn complexes | | Descriptor: | 5-(2-chlorophenyl)-N-hydroxy-1,3-oxazole-2-carboxamide, MANGANESE (II) ION, METHIONINE AMINOPEPTIDASE | | Authors: | Huguet, F, Melet, A, AlvesdeSousa, R, Lieutaud, A, Chevalier, J, Deschamps, P, Tomas, A, Leulliot, N, Pages, J.M, Artaud, I. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Hydroxamic Acids as Potent Inhibitors of Fe(II) and Mn(II) E. Coli Methionine Aminopeptidase: Biological Activities and X-Ray Structures of Oxazole Hydroxamate-Ecmetap-Mn Complexes.
Chemmedchem, 7, 2012
|
|
4A6V
 
 | | X-ray structures of oxazole hydroxamate EcMetAp-Mn complexes | | Descriptor: | CARBONATE ION, MANGANESE (II) ION, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Huguet, F, Melet, A, AlvesdeSousa, R, Lieutaud, A, Chevalier, J, Deschamps, P, Tomas, A, Leulliot, N, Pages, J.M, Artaud, I. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Hydroxamic Acids as Potent Inhibitors of Fe(II) and Mn(II) E. Coli Methionine Aminopeptidase: Biological Activities and X-Ray Structures of Oxazole Hydroxamate-Ecmetap-Mn Complexes.
Chemmedchem, 7, 2012
|
|
4C7P
 
 | | Crystal structure of Legionella pneumophila RalF F255K mutant | | Descriptor: | GLYCEROL, RALF | | Authors: | Folly-Klan, M, Alix, E, Stalder, D, Ray, P, Duarte, L.V, Delprato, A, Zeghouf, M, Antonny, B, Campanacci, V, Roy, C.R, Cherfils, J. | | Deposit date: | 2013-09-24 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Novel Membrane Sensor Controls the Localization and Arfgef Activity of Bacterial Ralf.
Plos Pathog., 9, 2013
|
|
4CBM
 
 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
6CAE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with NOSO-95179 antibiotic and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pantel, L, Florin, T, Dobosz-Bartoszek, M, Racine, E, Sarciaux, M, Serri, M, Houard, J, Campagne, J.M, Marcia de Figueiredo, R, Midrier, C, Gaudriault, S, Givaudan, A, Lanois, A, Forst, S, Aumelas, A, Cotteaux-Lautard, C, Bolla, J.M, Vingsbo Lundberg, C, Huseby, D, Hughes, D, Villain-Guillot, P, Mankin, A.S, Polikanov, Y.S, Gualtieri, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Odilorhabdins, Antibacterial Agents that Cause Miscoding by Binding at a New Ribosomal Site.
Mol. Cell, 70, 2018
|
|
6BHL
 
 | | Phosphotriesterase variant S5deltaL7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-10-30 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphotriesterase variant S5deltaL7
To Be Published
|
|
6D5Y
 
 | | Crystal structure of ERK2 G169D mutant | | Descriptor: | Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Jaiswal, B.S, Wang, W. | | Deposit date: | 2018-04-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | ERK Mutations and Amplification Confer Resistance to ERK-Inhibitor Therapy.
Clin. Cancer Res., 24, 2018
|
|
6BH7
 
 | | Phosphotriesterase variant R18+254S | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-10-30 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphotriesterase variant R18+254S
To Be Published
|
|
6BHK
 
 | | Phosphotriesterase variant R18deltaL7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphotriesterase, ZINC ION | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-10-30 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphotriesterase variant R18deltaL7
To Be Published
|
|
4B91
 
 | | Crystal structure of truncated human CRMP-5 | | Descriptor: | DIHYDROPYRIMIDINASE-RELATED PROTEIN 5 | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2012-08-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights Into the Oligomerization of Crmps: Crystal Structure of Human Collapsin Response Mediator Protein 5.
J.Neurochem., 125, 2013
|
|
6BGG
 
 | | Solution NMR structures of the BRD3 ET domain in complex with a CHD4 peptide | | Descriptor: | Bromodomain-containing protein 3, CHD4 | | Authors: | Wai, D.C.C, Szyszka, T.N, Campbell, A.E, Kwong, C, Wilkinson-White, L, Silva, A.P.G, Low, J.K.K, Kwan, A.H, Gamsjaeger, R, Lu, B, Vakoc, C.R, Blobel, G.A, Mackay, J.P. | | Deposit date: | 2017-10-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The BRD3 ET domain recognizes a short peptide motif through a mechanism that is conserved across chromatin remodelers and transcriptional regulators.
J. Biol. Chem., 293, 2018
|
|
