7JW0
 
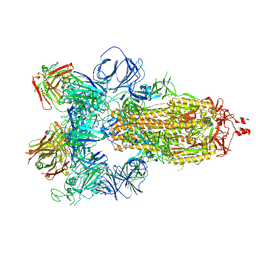 | | SARS-CoV-2 spike in complex with the S304 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S304 Fab heavy chain, ... | | Authors: | Walls, A.C, Park, Y.J, Tortorici, M.A, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
8T6I
 
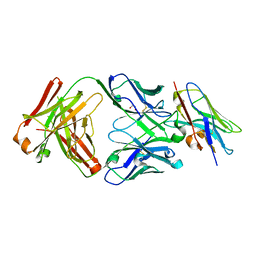 | | Structure of VHH-Fab complex with engineered Crystal Kappa region | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Filippova, E.V, Thompson, I, Kossiakoff, A.A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TS5
 
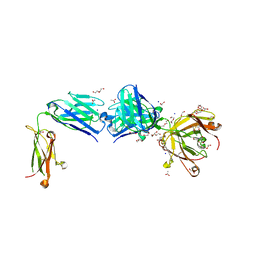 | | Structure of the apo FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L.L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TRT
 
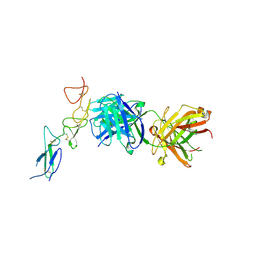 | | Structure of the EphA2 CRD bound to FabS1CE_C1, monoclinic form | | Descriptor: | CHLORIDE ION, Ephrin type-A receptor 2, S1CE variant of Fab C1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TRS
 
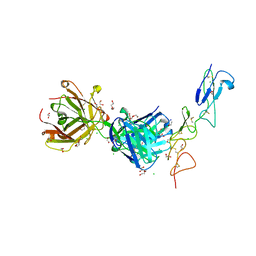 | | Structure of the EphA2 CRD bound to FabS1CE_C1, trigonal form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T8I
 
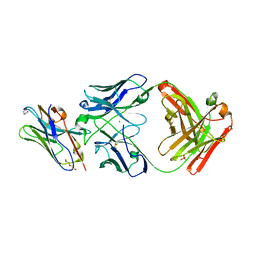 | | Structure of VHH-Fab complex with engineered Elbow FNQIKG, Crystal Kappa and SER substitutions | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fab heavy chain, ... | | Authors: | Filippova, E.V, Thompson, I, Kossiakoff, A.A. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T9Y
 
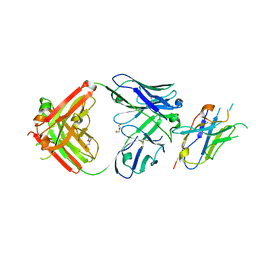 | | Structure of VHH-Fab complex with engineered Elbow FNQIKG and Crystal Kappa regions | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fab heavy chain, Fab light chain, ... | | Authors: | Filippova, E.V, Thompson, I, Kossiakoff, A.A. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
7U2D
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody ADG20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADG20 heavy chain, ADG20 light chain, ... | | Authors: | Zhu, X, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | A broad and potent neutralization epitope in SARS-related coronaviruses.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7JVC
 
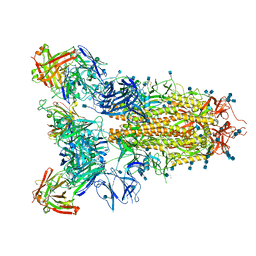 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2A4 Fab heavy chain, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
6PE7
 
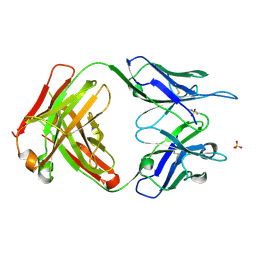 | | Crystal Structure of ABBV-323 FAB | | Descriptor: | FAB Heavy Chain, FAB Light chain, SULFATE ION | | Authors: | Argiriadi, M.A. | | Deposit date: | 2019-06-20 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | CD40/anti-CD40 antibody complexes which illustrate agonist and antagonist structural switches.
BMC Mol Cell Biol, 20, 2019
|
|
6PI7
 
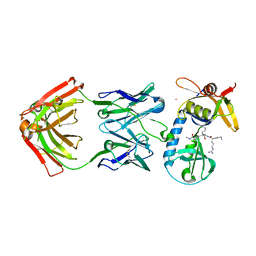 | |
6PE9
 
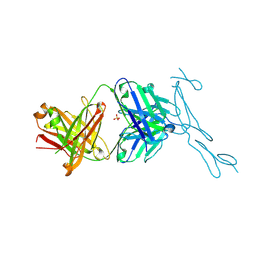 | | Crystal Structure of CD40 complexed to FAB516 | | Descriptor: | FAB Heavy chain, FAB Light chain, SULFATE ION, ... | | Authors: | Argiriadi, M.A. | | Deposit date: | 2019-06-20 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | CD40/anti-CD40 antibody complexes which illustrate agonist and antagonist structural switches.
BMC Mol Cell Biol, 20, 2019
|
|
6PE8
 
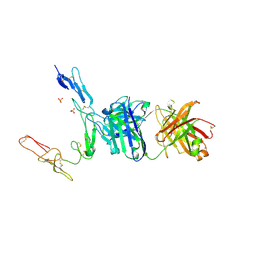 | | Crystal structure of CD40/ABBV-323 FAB complex | | Descriptor: | FAB Heavy chain, FAB Light chain, SULFATE ION, ... | | Authors: | Argiriadi, M.A. | | Deposit date: | 2019-06-20 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | CD40/anti-CD40 antibody complexes which illustrate agonist and antagonist structural switches.
BMC Mol Cell Biol, 20, 2019
|
|
4MWF
 
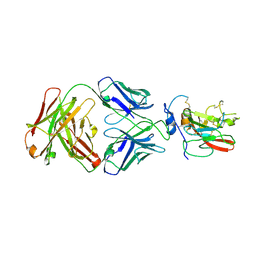 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core bound to broadly neutralizing antibody AR3C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Kong, L, Wilson, I.A, Law, M. | | Deposit date: | 2013-09-24 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Hepatitis C virus e2 envelope glycoprotein core structure.
Science, 342, 2013
|
|
4I1N
 
 | |
6S34
 
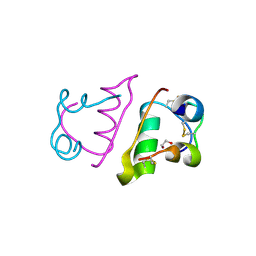 | |
8CWV
 
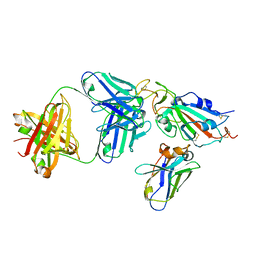 | |
7M6F
 
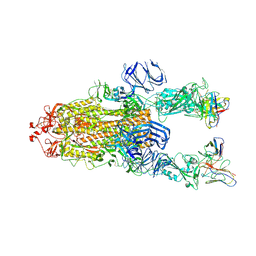 | |
7M6G
 
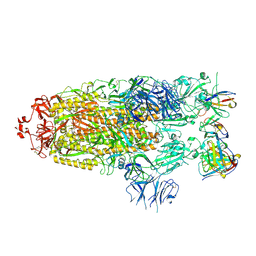 | |
7M6E
 
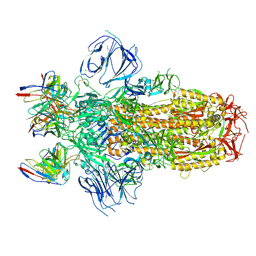 | |
8WO0
 
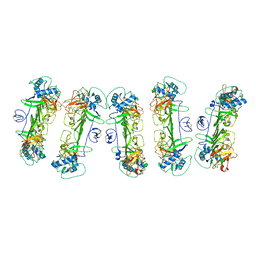 | | CryoEM structure of ZIKV rsNS1 filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Non-structural protein 1 | | Authors: | Chew, B.L.A, Luo, D. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis of Zika virus NS1 multimerization and human antibody recognition
Npj Viruses, 2, 2024
|
|
8WNU
 
 | | ZIKV rsNS1 in complex with Fab GB5 and anti-fab nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-fab nanobody, Non-structural protein 1, ... | | Authors: | Chew, B.L.A, Luo, D. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of Zika virus NS1 multimerization and human antibody recognition
Npj Viruses, 2, 2024
|
|
8WO4
 
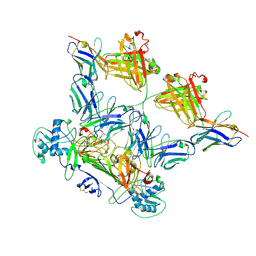 | | ZIKV rsNS1 in complex with Fab EB9 and anti-fab nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-fab nanobody, Non-structural protein 1, ... | | Authors: | Chew, B.L.A, Luo, D. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of Zika virus NS1 multimerization and human antibody recognition
Npj Viruses, 2, 2024
|
|
8WNP
 
 | | ZIKV rsNS1 in complex with Fab AA12 and anti-fab nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-fab nanobody, Non-structural protein 1, ... | | Authors: | Chew, B.L.A, Luo, D. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of Zika virus NS1 multimerization and human antibody recognition
Npj Viruses, 2, 2024
|
|
7Y58
 
 | |
