7GG0
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with PET-UNK-e44ffd04-1 (Mpro-x12026) | | Descriptor: | 2-(3-chlorophenyl)-N-[(4S)-[1,2,4]triazolo[4,3-a]pyridin-3-yl]acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.512 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHN
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-f2460aef-1 (Mpro-P0009) | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4AOJ
 
 | | Human TrkA in complex with the inhibitor AZ-23 | | Descriptor: | 5-chloranyl-N2-[(1S)-1-(5-fluoranylpyridin-2-yl)ethyl]-N4-(3-propan-2-yloxy-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, HIGH AFFINITY NERVE GROWTH FACTOR RECEPTOR, ZINC ION | | Authors: | Wang, T, Lamb, M.L, Block, M.H, Davies, A.M, Han, Y, Hoffmann, E, Ioannidis, S, Josey, J.A, Liu, Z, Lyne, P.D, MacIntyre, T, Mohr, P.J, Omer, C.A, Sjogren, T, Thress, K, Wang, B, Wang, H, Yu, D, Zhang, H. | | Deposit date: | 2012-03-28 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of Disubstituted Imidazo[4,5-B]Pyridines and Purines as Potent Trka Inhibitors
Acs Med.Chem.Lett., 3, 2012
|
|
3KQB
 
 | | Factor xa in complex with the inhibitor n-(3-fluoro-2'- (methylsulfonyl)biphenyl-4-yl)-1-(3-(5-oxo-4,5-dihydro-1h- 1,2,4-triazol-3-yl)phenyl)-3-(trifluoromethyl)-1h- pyrazole-5-carboxamide | | Descriptor: | N-(3-FLUORO-2'-(METHYLSULFONYL)BIPHENYL-4-YL)-1-(3-(5-OXO-4,5-DIHYDRO-1H-1,2,4-TRIAZOL-3-YL)PHENYL)-3-(TRIFLUOROMETHYL)-1H-PYRAZOLE-5-CARBOXAMIDE, SODIUM ION, factor Xa heavy chain, ... | | Authors: | Sheriff, S. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Phenyltriazolinones as potent factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7GGJ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BEN-DND-4f474d93-1 (Mpro-x12659) | | Descriptor: | (4R)-6-chloro-N-(2,7-naphthyridin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
3E6J
 
 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) RBC36 in Complex with H-trisaccharide | | Descriptor: | GLYCEROL, SULFATE ION, Variable lymphocyte receptor diversity region, ... | | Authors: | Han, B.W, Herrin, B.R, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Antigen recognition by variable lymphocyte receptors.
Science, 321, 2008
|
|
7GHP
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BEN-DND-4f474d93-1 (Mpro-P0012) | | Descriptor: | (4R)-6-chloro-N-(2,7-naphthyridin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5SE4
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(cc(nc2nc(nn12)CCc3nc(cn3C)c4ccccc4)C5CC5)C, micromolar IC50=0.001357 | | Descriptor: | (8S)-5-cyclopropyl-7-methyl-2-[2-(1-methyl-4-phenyl-1H-imidazol-2-yl)ethyl][1,2,4]triazolo[1,5-a]pyrimidine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SEH
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2(c(c(C(=O)N1CCC1)cn2)C(Nc4cc3nc(nn3cc4)N5CCOCC5)=O)C, micromolar IC50=0.002043 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[(4S)-2-(morpholin-4-yl)[1,2,4]triazolo[1,5-a]pyridin-7-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SPI
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z4574659604 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-1-{4-[(cyclopropanecarbonyl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-{4-[(cyclopropanecarbonyl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2C8H
 
 | | Structure of the PN loop Q182A mutant C3bot1 Exoenzyme (NAD-bound state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Properties of Wild-Type and Two Artt Motif Mutants Clostridium Botulinum C3 Exoenzyme Isoform 1 in Different Substrate Complexed States and Crystal Forms.
To be Published
|
|
6JXC
 
 | | Tel1 kinase butterfly symmetric dimer | | Descriptor: | Serine/threonine-protein kinase TEL1 | | Authors: | Xin, J. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of allosteric regulation of Tel1/ATM kinase.
Cell Res., 29, 2019
|
|
5SFB
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(cc(nn2c1nc(c2C)C)CCc3nc(nn3C)N4CCCC4)C(NC)=O, micromolar IC50=0.0021475 | | Descriptor: | (4S)-N,2,3-trimethyl-6-{2-[1-methyl-3-(pyrrolidin-1-yl)-1H-1,2,4-triazol-5-yl]ethyl}imidazo[1,2-b]pyridazine-8-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFH
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH N(C1CC1)(C)c2nn(c(n2)CCc4nn3c(cnc(c3n4)C)C)C, micromolar IC50=0.042255 | | Descriptor: | MAGNESIUM ION, N-cyclopropyl-5-{2-[(4S)-5,8-dimethyl[1,2,4]triazolo[1,5-a]pyrazin-2-yl]ethyl}-N,1-dimethyl-1H-1,2,4-triazol-3-amine, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
3KWK
 
 | |
6CC7
 
 | | Structure of Fungal GH62 from Thielavia terretris | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycoside hydrolase family 62 protein, ... | | Authors: | Camargo, S, Mulinari, E.J, Almeida, L.R, Muniz, J.R.C. | | Deposit date: | 2018-02-06 | | Release date: | 2019-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of Fungal GH62 from Thielavia terretris
To Be Published
|
|
456C
 
 | | CRYSTAL STRUCTURE OF COLLAGENASE-3 (MMP-13) COMPLEXED TO A DIPHENYL-ETHER SULPHONE BASED HYDROXAMIC ACID | | Descriptor: | 2-{4-[4-(4-CHLORO-PHENOXY)-BENZENESULFONYL]-TETRAHYDRO-PYRAN-4-YL}-N-HYDROXY-ACETAMIDE, CALCIUM ION, MMP-13, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
5E1S
 
 | | The Crystal structure of INSR Tyrosine Kinase in complex with the Inhibitor BI 885578 | | Descriptor: | (5R)-N-(1-{2-[4-(2-methoxyethyl)piperazin-1-yl]ethyl}-1H-pyrazol-3-yl)-5,8-dimethyl-9-phenyl-6,8-dihydro-5H-pyrazolo[3,4-h]quinazolin-2-amine, Insulin receptor | | Authors: | Kessler, D, Zahn, S, Sanderson, M, Wolkerstorfer, B. | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.264 Å) | | Cite: | BI 885578, a Novel IGF1R/INSR Tyrosine Kinase Inhibitor with Pharmacokinetic Properties That Dissociate Antitumor Efficacy and Perturbation of Glucose Homeostasis.
Mol.Cancer Ther., 14, 2015
|
|
4QGP
 
 | |
4QKX
 
 | | Structure of beta2 adrenoceptor bound to a covalent agonist and an engineered nanobody | | Descriptor: | 4-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(2-sulfanylethoxy)phenyl]ethyl}amino)ethyl]benzene-1,2-diol, Beta-2 adrenergic receptor, R9 protein, ... | | Authors: | Weichert, D, Kruse, A.C, Manglik, A, Hiller, C, Zhang, C, Huebner, H, Kobilka, B.K, Gmeiner, P. | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Covalent agonists for studying G protein-coupled receptor activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ITU
 
 | | hPDE2A catalytic domain complexed with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2009-08-28 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5YWX
 
 | | Crystal structure of hematopoietic prostaglandin D synthase in complex with F092 | | Descriptor: | GLUTATHIONE, GLYCEROL, Hematopoietic prostaglandin D synthase, ... | | Authors: | Kamo, M, Furubayashi, N, Inaka, K, Aritake, K, Omura, A, Tanaka, A. | | Deposit date: | 2017-11-30 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Characterization of crystal water molecules in a high-affinity inhibitor and hematopoietic prostaglandin D synthase complex by interaction energy studies.
Bioorg. Med. Chem., 26, 2018
|
|
3IWE
 
 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD85646 | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD85646
To be Published
|
|
6MZI
 
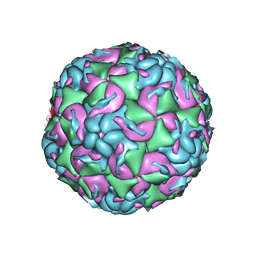 | | CryoEM structure of human enterovirus D68 expanded 1 particle (pH 6.5, 4 degrees Celsius, 3 min) | | Descriptor: | viral protein 1, viral protein 2, viral protein 3, ... | | Authors: | Liu, Y, Rossmann, M.G. | | Deposit date: | 2018-11-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Molecular basis for the acid-initiated uncoating of human enterovirus D68.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Z0Y
 
 | |
